Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2412 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2412 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 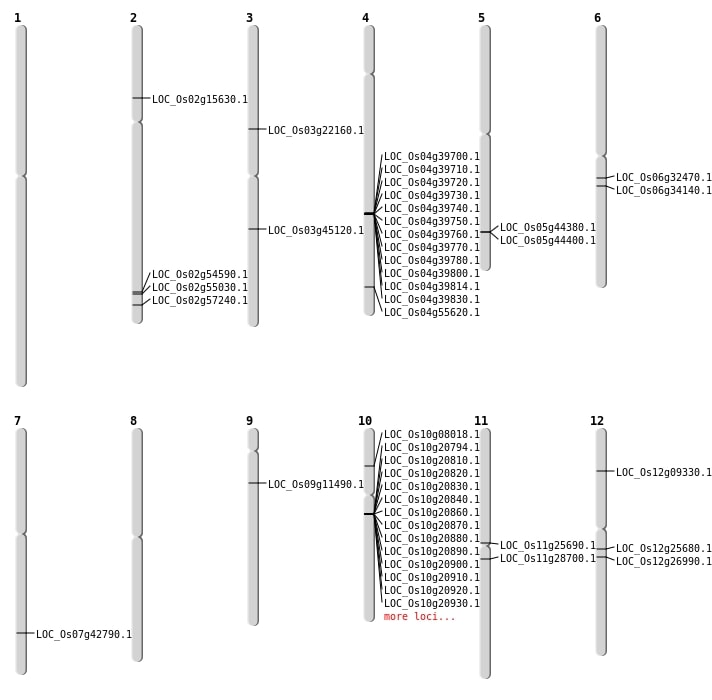
|
| Variant Information |
|---|
Single base substitutions: 58
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 19754295 | G-A | HET | ||
| SBS | Chr1 | 23003215 | G-C | HET | ||
| SBS | Chr1 | 23679422 | G-T | HET | ||
| SBS | Chr1 | 24762649 | G-A | HET | ||
| SBS | Chr1 | 36547446 | C-T | HET | ||
| SBS | Chr10 | 13461896 | C-A | HET | ||
| SBS | Chr10 | 15159146 | T-C | HET | ||
| SBS | Chr10 | 16688162 | A-G | Homo | ||
| SBS | Chr10 | 4340128 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os10g08018 |
| SBS | Chr11 | 14653027 | C-A | Homo | STOP_GAINED | LOC_Os11g25690 |
| SBS | Chr11 | 16593829 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os11g28700 |
| SBS | Chr11 | 25042746 | G-A | HET | ||
| SBS | Chr12 | 14386291 | C-A | Homo | ||
| SBS | Chr12 | 14883421 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g25680 |
| SBS | Chr12 | 15810132 | T-C | Homo | NON_SYNONYMOUS_CODING | LOC_Os12g26990 |
| SBS | Chr12 | 15989418 | A-T | Homo | ||
| SBS | Chr12 | 4896610 | A-C | Homo | NON_SYNONYMOUS_CODING | LOC_Os12g09330 |
| SBS | Chr12 | 5651549 | G-A | HET | ||
| SBS | Chr2 | 12400378 | T-C | HET | ||
| SBS | Chr2 | 22267407 | A-T | Homo | ||
| SBS | Chr2 | 28196743 | C-T | Homo | ||
| SBS | Chr2 | 35068149 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os02g57240 |
| SBS | Chr2 | 6216164 | G-A | HET | ||
| SBS | Chr3 | 10384604 | C-T | HET | ||
| SBS | Chr3 | 13917715 | A-G | HET | ||
| SBS | Chr3 | 18508502 | G-T | HET | ||
| SBS | Chr3 | 20055353 | G-T | Homo | ||
| SBS | Chr3 | 25464291 | C-T | HET | SPLICE_SITE_ACCEPTOR | LOC_Os03g45120 |
| SBS | Chr3 | 26934120 | C-T | HET | ||
| SBS | Chr3 | 426375 | T-G | HET | ||
| SBS | Chr4 | 10145304 | C-A | HET | ||
| SBS | Chr4 | 14577948 | G-C | HET | ||
| SBS | Chr4 | 17123845 | G-A | HET | ||
| SBS | Chr5 | 13605367 | A-T | HET | ||
| SBS | Chr5 | 13605368 | A-T | HET | ||
| SBS | Chr5 | 13850590 | C-T | HET | ||
| SBS | Chr5 | 14061090 | C-T | HET | ||
| SBS | Chr5 | 20671160 | C-A | HET | ||
| SBS | Chr5 | 23319220 | G-A | HET | ||
| SBS | Chr5 | 23896995 | T-G | HET | ||
| SBS | Chr5 | 24752691 | G-A | HET | ||
| SBS | Chr5 | 9551436 | G-A | HET | ||
| SBS | Chr6 | 10212257 | T-C | HET | ||
| SBS | Chr6 | 11031128 | A-T | Homo | ||
| SBS | Chr6 | 11497607 | T-C | Homo | ||
| SBS | Chr6 | 16129979 | C-A | HET | ||
| SBS | Chr6 | 22412206 | A-T | HET | ||
| SBS | Chr6 | 2596531 | G-A | HET | ||
| SBS | Chr6 | 3595079 | T-C | HET | ||
| SBS | Chr6 | 7907989 | T-G | HET | ||
| SBS | Chr7 | 26969619 | T-C | Homo | ||
| SBS | Chr7 | 3543856 | C-A | Homo | ||
| SBS | Chr8 | 19135584 | G-A | HET | ||
| SBS | Chr8 | 5550492 | T-C | HET | ||
| SBS | Chr9 | 14230785 | G-A | HET | ||
| SBS | Chr9 | 3222801 | A-G | HET | ||
| SBS | Chr9 | 6412666 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os09g11490 |
| SBS | Chr9 | 8291974 | A-T | HET |
Deletions: 16
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr5 | 3337621 | 3337627 | 6 | |
| Deletion | Chr12 | 5274832 | 5274837 | 5 | |
| Deletion | Chr10 | 10544001 | 10608000 | 63999 | 16 |
| Deletion | Chr8 | 11800580 | 11800582 | 2 | |
| Deletion | Chr3 | 12694001 | 12708000 | 13999 | LOC_Os03g22160 |
| Deletion | Chr9 | 15663691 | 15663693 | 2 | |
| Deletion | Chr8 | 17058106 | 17058156 | 50 | |
| Deletion | Chr6 | 19886304 | 19886308 | 4 | LOC_Os06g34140 |
| Deletion | Chr5 | 20080234 | 20080236 | 2 | |
| Deletion | Chr3 | 20402302 | 20402303 | 1 | |
| Deletion | Chr4 | 23660001 | 23675000 | 14999 | 4 |
| Deletion | Chr4 | 23684001 | 23726000 | 41999 | 8 |
| Deletion | Chr12 | 24394714 | 24394718 | 4 | |
| Deletion | Chr4 | 28682998 | 28682999 | 1 | |
| Deletion | Chr6 | 30975185 | 30975200 | 15 | |
| Deletion | Chr4 | 34712012 | 34712023 | 11 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr10 | 3386611 | 3386612 | 2 | |
| Insertion | Chr11 | 18215515 | 18215515 | 1 | |
| Insertion | Chr5 | 10007414 | 10007414 | 1 |
Inversions: 7
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr8 | 10782911 | 10783808 | |
| Inversion | Chr12 | 15810552 | 16085304 | LOC_Os12g26990 |
| Inversion | Chr12 | 24899750 | 25423356 | |
| Inversion | Chr12 | 24899756 | 25423361 | |
| Inversion | Chr7 | 25633734 | 26209881 | LOC_Os07g42790 |
| Inversion | Chr2 | 33692648 | 33716503 | 2 |
| Inversion | Chr2 | 33692734 | 33716510 | 2 |
Translocations: 9
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr8 | 10668730 | Chr6 | 18881069 | 2 |
| Translocation | Chr8 | 10778209 | Chr7 | 25633744 | 2 |
| Translocation | Chr8 | 10782915 | Chr7 | 26210343 | |
| Translocation | Chr5 | 25821980 | Chr2 | 33436575 | 2 |
| Translocation | Chr5 | 25834120 | Chr2 | 33437748 | 2 |
| Translocation | Chr7 | 26209883 | Chr4 | 2784322 | |
| Translocation | Chr7 | 26210333 | Chr4 | 2784320 | |
| Translocation | Chr11 | 28627092 | Chr10 | 752826 | |
| Translocation | Chr4 | 33111949 | Chr2 | 8784357 | 2 |
