Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN244-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN244-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 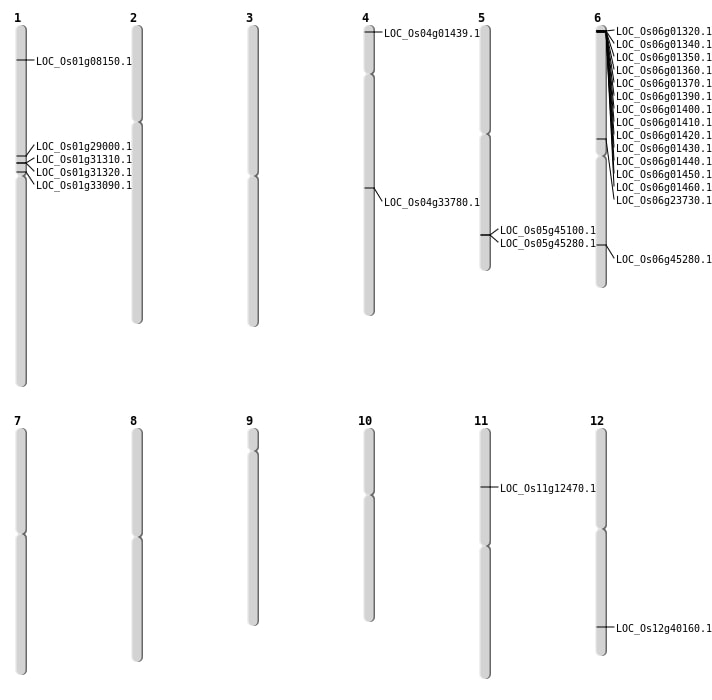
|
| Variant Information |
|---|
Single base substitutions: 35
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 13709425 | A-T | HET | INTERGENIC | |
| SBS | Chr1 | 16236597 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g29000 |
| SBS | Chr1 | 24958491 | G-C | HOMO | INTERGENIC | |
| SBS | Chr1 | 29026017 | A-G | HOMO | INTERGENIC | |
| SBS | Chr1 | 39221662 | T-G | HET | INTERGENIC | |
| SBS | Chr1 | 39262332 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 8467909 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 2337941 | A-T | HET | INTERGENIC | |
| SBS | Chr10 | 4934277 | C-A | HET | INTERGENIC | |
| SBS | Chr10 | 6183740 | C-G | HET | INTERGENIC | |
| SBS | Chr10 | 7794856 | G-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 24321010 | C-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 7081471 | A-G | HET | INTERGENIC | |
| SBS | Chr12 | 18793399 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 5471417 | C-T | HET | INTRON | |
| SBS | Chr3 | 15822283 | C-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 22034501 | C-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 4168990 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 304714 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os04g01439 |
| SBS | Chr5 | 14292249 | G-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 15602732 | G-T | HET | INTERGENIC | |
| SBS | Chr5 | 15602733 | T-G | HET | INTERGENIC | |
| SBS | Chr5 | 15602735 | G-T | HET | INTERGENIC | |
| SBS | Chr5 | 20964280 | T-C | HET | INTERGENIC | |
| SBS | Chr5 | 20964281 | A-G | HET | INTERGENIC | |
| SBS | Chr5 | 26198853 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g45100 |
| SBS | Chr5 | 29781798 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 13868968 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os06g23730 |
| SBS | Chr6 | 14017837 | A-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr6 | 14929312 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 22103500 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 8570145 | C-A | HET | INTERGENIC | |
| SBS | Chr8 | 13298 | G-T | HOMO | INTRON | |
| SBS | Chr8 | 863209 | T-A | HET | INTERGENIC | |
| SBS | Chr8 | 9931226 | C-T | HET | INTERGENIC |
Deletions: 24
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr6 | 201621 | 276983 | 75362 | 13 |
| Deletion | Chr7 | 1574002 | 1574005 | 3 | |
| Deletion | Chr1 | 3942243 | 3942251 | 8 | LOC_Os01g08150 |
| Deletion | Chr1 | 5528656 | 5528672 | 16 | |
| Deletion | Chr3 | 5720334 | 5720339 | 5 | |
| Deletion | Chr10 | 5865882 | 5865889 | 7 | |
| Deletion | Chr10 | 7703710 | 7703715 | 5 | |
| Deletion | Chr1 | 10759902 | 10759922 | 20 | |
| Deletion | Chr5 | 11356667 | 11356679 | 12 | |
| Deletion | Chr5 | 11363885 | 11363888 | 3 | |
| Deletion | Chr5 | 13913215 | 13913222 | 7 | |
| Deletion | Chr9 | 15955234 | 15955236 | 2 | |
| Deletion | Chr10 | 16071135 | 16071139 | 4 | |
| Deletion | Chr11 | 17631096 | 17631123 | 27 | |
| Deletion | Chr2 | 19222109 | 19222114 | 5 | |
| Deletion | Chr4 | 20462730 | 20462739 | 9 | LOC_Os04g33780 |
| Deletion | Chr3 | 22960615 | 22960630 | 15 | |
| Deletion | Chr1 | 23389536 | 23389547 | 11 | |
| Deletion | Chr12 | 23554923 | 23554925 | 2 | |
| Deletion | Chr12 | 24857388 | 24857402 | 14 | LOC_Os12g40160 |
| Deletion | Chr11 | 27150495 | 27150497 | 2 | |
| Deletion | Chr6 | 27369158 | 27369162 | 4 | LOC_Os06g45280 |
| Deletion | Chr6 | 28410834 | 28410835 | 1 | |
| Deletion | Chr4 | 31813302 | 31813320 | 18 |
Insertions: 9
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr11 | 6969230 | 7083645 | LOC_Os11g12470 |
| Inversion | Chr11 | 6978292 | 7083653 | |
| Inversion | Chr1 | 17136302 | 18206821 | 2 |
| Inversion | Chr1 | 17137262 | 18206829 | 2 |
| Inversion | Chr5 | 26282072 | 27040358 | LOC_Os05g45280 |
No Translocation
