Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2440 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2440 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 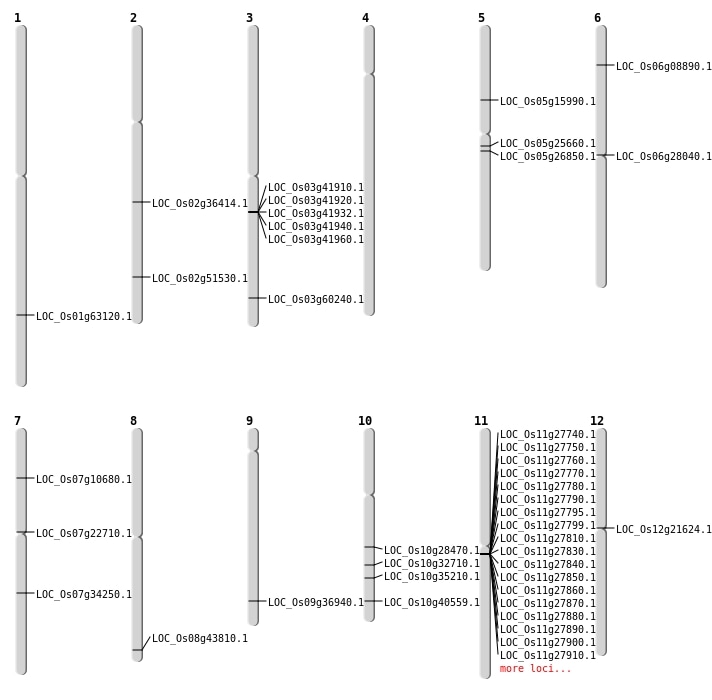
|
| Variant Information |
|---|
Single base substitutions: 57
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 11636794 | G-C | HET | ||
| SBS | Chr1 | 11764392 | C-A | HET | ||
| SBS | Chr1 | 29188520 | C-T | HET | ||
| SBS | Chr1 | 36578099 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g63120 |
| SBS | Chr1 | 39153960 | A-T | HET | ||
| SBS | Chr1 | 6016343 | G-T | HET | ||
| SBS | Chr1 | 7995128 | C-T | HET | ||
| SBS | Chr10 | 12473061 | T-C | HET | ||
| SBS | Chr10 | 14813865 | G-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os10g28470 |
| SBS | Chr10 | 16588864 | T-C | Homo | ||
| SBS | Chr10 | 18809866 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os10g35210 |
| SBS | Chr10 | 20588353 | C-T | HET | ||
| SBS | Chr11 | 15241299 | A-G | HET | ||
| SBS | Chr11 | 23000730 | A-C | Homo | ||
| SBS | Chr11 | 24877559 | T-C | Homo | ||
| SBS | Chr11 | 26479678 | T-A | Homo | ||
| SBS | Chr11 | 28681436 | A-G | HET | ||
| SBS | Chr11 | 4690594 | G-T | HET | ||
| SBS | Chr11 | 8374860 | A-G | HET | ||
| SBS | Chr2 | 16834154 | T-C | HET | ||
| SBS | Chr2 | 31485965 | T-C | HET | ||
| SBS | Chr2 | 31559558 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os02g51530 |
| SBS | Chr2 | 3214623 | T-C | HET | ||
| SBS | Chr2 | 34396684 | C-A | HET | ||
| SBS | Chr3 | 10384718 | A-C | HET | ||
| SBS | Chr3 | 11651335 | C-T | Homo | ||
| SBS | Chr3 | 15162015 | T-A | Homo | ||
| SBS | Chr3 | 23780685 | G-A | HET | ||
| SBS | Chr3 | 3202182 | T-A | HET | ||
| SBS | Chr4 | 10108036 | C-T | HET | ||
| SBS | Chr4 | 19829288 | G-A | HET | ||
| SBS | Chr4 | 22612168 | A-C | HET | ||
| SBS | Chr4 | 23281883 | G-A | HET | ||
| SBS | Chr4 | 8637078 | G-A | HET | ||
| SBS | Chr5 | 10594627 | T-G | HET | ||
| SBS | Chr5 | 15301639 | G-A | HET | ||
| SBS | Chr5 | 1914928 | A-G | HET | ||
| SBS | Chr5 | 21725010 | C-T | HET | ||
| SBS | Chr5 | 237660 | G-T | HET | ||
| SBS | Chr5 | 26775023 | A-T | HET | ||
| SBS | Chr5 | 6508760 | G-A | HET | ||
| SBS | Chr5 | 9026728 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os05g15990 |
| SBS | Chr5 | 9285883 | C-A | Homo | ||
| SBS | Chr6 | 11232197 | G-T | HET | ||
| SBS | Chr6 | 13885815 | G-A | HET | ||
| SBS | Chr6 | 30943596 | T-A | HET | ||
| SBS | Chr6 | 31060911 | C-T | HET | ||
| SBS | Chr6 | 4473539 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os06g08890 |
| SBS | Chr6 | 8136425 | G-A | HET | ||
| SBS | Chr7 | 10473080 | T-G | HET | ||
| SBS | Chr7 | 3187676 | C-T | Homo | ||
| SBS | Chr7 | 8472906 | T-G | Homo | ||
| SBS | Chr8 | 11068783 | C-T | Homo | ||
| SBS | Chr8 | 27655991 | T-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os08g43810 |
| SBS | Chr8 | 5394581 | C-T | Homo | ||
| SBS | Chr9 | 16451398 | C-T | HET | ||
| SBS | Chr9 | 4261660 | C-A | HET |
Deletions: 23
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr6 | 5084934 | 5084946 | 12 | |
| Deletion | Chr2 | 5274764 | 5274770 | 6 | |
| Deletion | Chr4 | 7501459 | 7501472 | 13 | |
| Deletion | Chr2 | 10756220 | 10756221 | 1 | |
| Deletion | Chr12 | 12150956 | 12150964 | 8 | LOC_Os12g21624 |
| Deletion | Chr7 | 12793001 | 12808000 | 14999 | LOC_Os07g22710 |
| Deletion | Chr7 | 12811227 | 12811228 | 1 | |
| Deletion | Chr1 | 14031736 | 14031753 | 17 | |
| Deletion | Chr9 | 14322247 | 14322252 | 5 | |
| Deletion | Chr2 | 15443029 | 15443033 | 4 | |
| Deletion | Chr11 | 15969001 | 16175000 | 205999 | 37 |
| Deletion | Chr11 | 16181001 | 16207000 | 25999 | 3 |
| Deletion | Chr10 | 17124403 | 17124412 | 9 | LOC_Os10g32710 |
| Deletion | Chr9 | 21313569 | 21313572 | 3 | LOC_Os09g36940 |
| Deletion | Chr10 | 21722210 | 21722212 | 2 | LOC_Os10g40559 |
| Deletion | Chr2 | 21995626 | 21995627 | 1 | LOC_Os02g36414 |
| Deletion | Chr3 | 23269001 | 23306000 | 36999 | 5 |
| Deletion | Chr11 | 24808462 | 24808463 | 1 | |
| Deletion | Chr5 | 25624708 | 25624711 | 3 | |
| Deletion | Chr8 | 27656283 | 27656284 | 1 | LOC_Os08g43810 |
| Deletion | Chr4 | 28169102 | 28169103 | 1 | |
| Deletion | Chr4 | 31876511 | 31876512 | 1 | |
| Deletion | Chr3 | 34261916 | 34261922 | 6 | LOC_Os03g60240 |
Insertions: 6
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr5 | 14921582 | 15910410 | LOC_Os05g25660 |
| Inversion | Chr5 | 15902290 | 15903203 | |
| Inversion | Chr5 | 15903205 | 16215708 | |
| Inversion | Chr7 | 20337208 | 20502930 | 2 |
Translocations: 6
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr7 | 5800924 | Chr3 | 24163935 | LOC_Os07g10680 |
| Translocation | Chr7 | 5800927 | Chr3 | 24165770 | LOC_Os07g10680 |
| Translocation | Chr5 | 15586230 | Chr1 | 39146568 | LOC_Os05g26850 |
| Translocation | Chr6 | 15911333 | Chr5 | 4678636 | LOC_Os06g28040 |
| Translocation | Chr11 | 17285418 | Chr5 | 14921559 | 2 |
| Translocation | Chr11 | 17285440 | Chr5 | 14920630 |
