Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2456 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2456 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 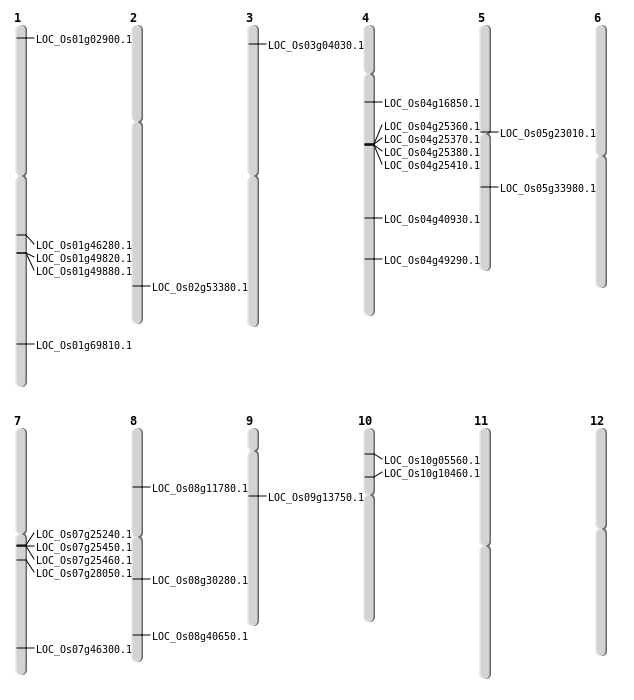
|
| Variant Information |
|---|
Single base substitutions: 46
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 1057849 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g02900 |
| SBS | Chr1 | 13405165 | T-C | HET | ||
| SBS | Chr1 | 2680370 | C-T | HET | ||
| SBS | Chr1 | 28254768 | C-G | HET | ||
| SBS | Chr11 | 14480752 | G-A | Homo | ||
| SBS | Chr11 | 16793691 | G-A | HET | ||
| SBS | Chr11 | 6236128 | C-A | HET | ||
| SBS | Chr12 | 15709312 | C-T | HET | ||
| SBS | Chr12 | 16899725 | T-C | HET | ||
| SBS | Chr12 | 5511820 | C-T | HET | ||
| SBS | Chr12 | 8261037 | T-C | HET | ||
| SBS | Chr2 | 18310657 | G-A | Homo | ||
| SBS | Chr2 | 19150315 | A-T | Homo | ||
| SBS | Chr2 | 32673594 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os02g53380 |
| SBS | Chr2 | 34741461 | G-T | HET | ||
| SBS | Chr2 | 7516399 | A-G | HET | ||
| SBS | Chr3 | 13171279 | A-T | HET | ||
| SBS | Chr3 | 17825902 | T-G | HET | ||
| SBS | Chr3 | 1838424 | C-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os03g04030 |
| SBS | Chr3 | 19208480 | G-A | HET | ||
| SBS | Chr3 | 25006039 | G-T | HET | ||
| SBS | Chr3 | 31965819 | T-C | HET | ||
| SBS | Chr3 | 8393856 | G-C | HET | ||
| SBS | Chr4 | 19045630 | C-T | Homo | ||
| SBS | Chr4 | 20613479 | T-C | Homo | ||
| SBS | Chr4 | 24300409 | C-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os04g40930 |
| SBS | Chr4 | 28908060 | T-C | Homo | ||
| SBS | Chr4 | 30644821 | A-T | HET | ||
| SBS | Chr4 | 9224469 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g16850 |
| SBS | Chr5 | 13876903 | G-A | HET | ||
| SBS | Chr5 | 14880853 | G-A | HET | ||
| SBS | Chr5 | 20063974 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g33980 |
| SBS | Chr5 | 20369899 | T-A | HET | ||
| SBS | Chr6 | 1281097 | G-A | HET | ||
| SBS | Chr6 | 25778143 | G-A | Homo | ||
| SBS | Chr7 | 14426843 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g25240 |
| SBS | Chr7 | 18745419 | G-T | HET | ||
| SBS | Chr7 | 22718930 | T-C | HET | ||
| SBS | Chr7 | 22788045 | C-T | HET | ||
| SBS | Chr7 | 2395979 | G-A | Homo | ||
| SBS | Chr7 | 24413735 | A-T | HET | ||
| SBS | Chr7 | 26710332 | G-T | HET | ||
| SBS | Chr7 | 27623248 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os07g46300 |
| SBS | Chr8 | 10148371 | C-A | HET | ||
| SBS | Chr8 | 6901763 | T-G | HET | NON_SYNONYMOUS_CODING | LOC_Os08g11780 |
| SBS | Chr9 | 20564719 | T-A | HET |
Deletions: 9
Insertions: 5
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 40308970 | 40308970 | 1 | LOC_Os01g69810 |
| Insertion | Chr11 | 26126640 | 26126640 | 1 | |
| Insertion | Chr3 | 11236907 | 11236912 | 6 | |
| Insertion | Chr8 | 16899200 | 16899205 | 6 | |
| Insertion | Chr9 | 8057872 | 8057872 | 1 | LOC_Os09g13750 |
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr7 | 20942096 | 21295095 | |
| Inversion | Chr7 | 20942106 | 21295107 | |
| Inversion | Chr1 | 28607668 | 28651372 | 2 |
| Inversion | Chr1 | 28607680 | 28651378 | 2 |
Translocations: 10
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr10 | 2761235 | Chr1 | 26316013 | 2 |
| Translocation | Chr9 | 5120391 | Chr4 | 3943179 | |
| Translocation | Chr10 | 5747369 | Chr5 | 13127961 | 2 |
| Translocation | Chr9 | 9765905 | Chr2 | 6850978 | |
| Translocation | Chr9 | 10309860 | Chr4 | 1632054 | |
| Translocation | Chr7 | 11077830 | Chr4 | 29420496 | 2 |
| Translocation | Chr7 | 13890993 | Chr4 | 15170551 | |
| Translocation | Chr8 | 18631189 | Chr7 | 16354719 | 2 |
| Translocation | Chr7 | 22771291 | Chr6 | 14580538 | |
| Translocation | Chr11 | 28719466 | Chr8 | 25742774 | 2 |
