Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2462 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2462 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 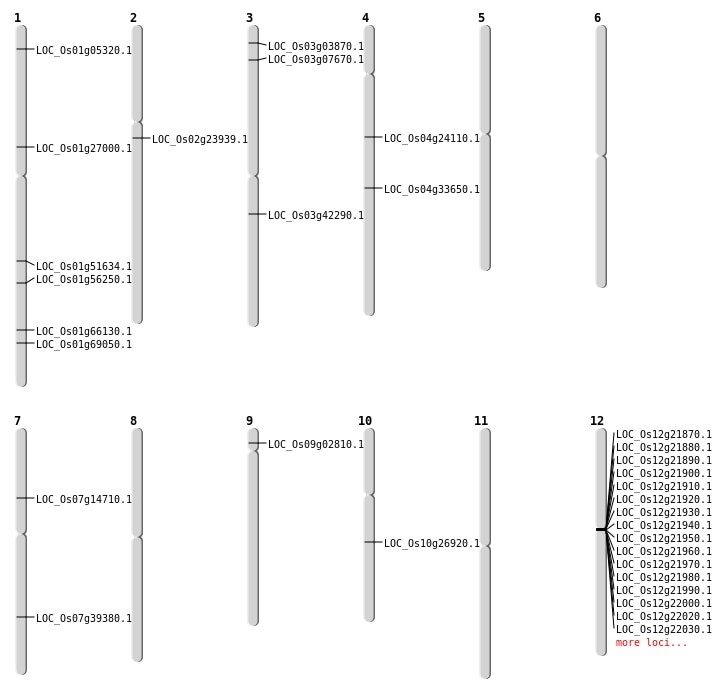
|
| Variant Information |
|---|
Single base substitutions: 54
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 15043653 | C-G | Homo | NON_SYNONYMOUS_CODING | LOC_Os01g27000 |
| SBS | Chr1 | 29673077 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g51634 |
| SBS | Chr1 | 32395790 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g56250 |
| SBS | Chr10 | 13654772 | G-T | HET | ||
| SBS | Chr10 | 14159108 | C-T | Homo | ||
| SBS | Chr10 | 14159109 | A-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os10g26920 |
| SBS | Chr10 | 2684557 | T-A | HET | ||
| SBS | Chr10 | 9661047 | T-A | HET | ||
| SBS | Chr11 | 10164562 | C-A | HET | ||
| SBS | Chr11 | 15242534 | C-T | Homo | ||
| SBS | Chr11 | 17789173 | A-T | HET | ||
| SBS | Chr11 | 18767699 | C-T | Homo | ||
| SBS | Chr11 | 873394 | T-C | HET | ||
| SBS | Chr12 | 14585014 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os12g25320 |
| SBS | Chr12 | 15582537 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os12g26610 |
| SBS | Chr12 | 27422282 | C-A | HET | ||
| SBS | Chr2 | 1178242 | G-T | HET | ||
| SBS | Chr2 | 13591144 | C-T | HET | ||
| SBS | Chr2 | 13867335 | C-T | HET | STOP_GAINED | LOC_Os02g23939 |
| SBS | Chr2 | 18540807 | C-T | HET | ||
| SBS | Chr2 | 23195903 | G-A | HET | ||
| SBS | Chr2 | 29830543 | C-T | HET | ||
| SBS | Chr2 | 31044923 | C-T | HET | ||
| SBS | Chr2 | 7637410 | G-A | HET | ||
| SBS | Chr3 | 1757169 | C-T | HET | STOP_GAINED | LOC_Os03g03870 |
| SBS | Chr3 | 1757170 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g03870 |
| SBS | Chr3 | 2082047 | T-G | HET | ||
| SBS | Chr3 | 23537559 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g42290 |
| SBS | Chr3 | 28866596 | A-T | Homo | ||
| SBS | Chr4 | 11149944 | T-A | Homo | ||
| SBS | Chr4 | 13771712 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g24110 |
| SBS | Chr4 | 15128357 | C-A | HET | ||
| SBS | Chr4 | 20381031 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g33650 |
| SBS | Chr4 | 7839501 | C-T | HET | ||
| SBS | Chr5 | 12062597 | G-A | Homo | ||
| SBS | Chr5 | 12136281 | G-T | Homo | ||
| SBS | Chr5 | 24773516 | C-G | HET | ||
| SBS | Chr5 | 29750809 | G-A | Homo | ||
| SBS | Chr5 | 9163394 | G-T | HET | ||
| SBS | Chr6 | 12605210 | C-A | HET | ||
| SBS | Chr6 | 20446344 | T-C | HET | ||
| SBS | Chr6 | 21426890 | A-G | HET | ||
| SBS | Chr6 | 5752925 | A-G | HET | ||
| SBS | Chr7 | 10473927 | T-A | HET | ||
| SBS | Chr7 | 21888529 | T-A | HET | ||
| SBS | Chr7 | 22807200 | A-T | HET | ||
| SBS | Chr7 | 23591489 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g39380 |
| SBS | Chr7 | 8391781 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os07g14710 |
| SBS | Chr8 | 15818673 | C-G | HET | ||
| SBS | Chr8 | 24786560 | C-A | HET | ||
| SBS | Chr8 | 4724644 | A-G | HET | ||
| SBS | Chr8 | 6333852 | G-T | HET | ||
| SBS | Chr8 | 8464265 | G-A | HET | ||
| SBS | Chr9 | 15257859 | C-G | HET |
Deletions: 19
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr1 | 2507302 | 2507304 | 2 | LOC_Os01g05320 |
| Deletion | Chr5 | 4617853 | 4617854 | 1 | |
| Deletion | Chr11 | 9444955 | 9444968 | 13 | |
| Deletion | Chr9 | 12225283 | 12225284 | 1 | |
| Deletion | Chr12 | 12316001 | 12539000 | 222999 | 37 |
| Deletion | Chr12 | 12875001 | 13001000 | 125999 | 17 |
| Deletion | Chr12 | 13004001 | 13051000 | 46999 | 6 |
| Deletion | Chr12 | 13054001 | 13134000 | 79999 | 15 |
| Deletion | Chr12 | 13154001 | 13339000 | 184999 | 27 |
| Deletion | Chr12 | 13424001 | 13463000 | 38999 | 8 |
| Deletion | Chr4 | 14237929 | 14237930 | 1 | |
| Deletion | Chr3 | 14330610 | 14330622 | 12 | |
| Deletion | Chr11 | 16289665 | 16289668 | 3 | |
| Deletion | Chr11 | 17108731 | 17108732 | 1 | |
| Deletion | Chr11 | 24651582 | 24651587 | 5 | |
| Deletion | Chr6 | 27804227 | 27804234 | 7 | |
| Deletion | Chr1 | 38412441 | 38412444 | 3 | LOC_Os01g66130 |
| Deletion | Chr1 | 40131960 | 40131963 | 3 | LOC_Os01g69050 |
| Deletion | Chr1 | 40684890 | 40684899 | 9 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr11 | 368917 | 368917 | 1 | |
| Insertion | Chr3 | 3895807 | 3895807 | 1 | LOC_Os03g07670 |
| Insertion | Chr7 | 17560179 | 17560180 | 2 |
No Inversion
Translocations: 8
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 1298238 | Chr7 | 19657642 | |
| Translocation | Chr9 | 1310136 | Chr8 | 2941961 | LOC_Os09g02810 |
| Translocation | Chr9 | 1311709 | Chr8 | 2941966 | LOC_Os09g02810 |
| Translocation | Chr12 | 10174306 | Chr4 | 8429189 | |
| Translocation | Chr12 | 14705801 | Chr3 | 8253738 | |
| Translocation | Chr9 | 15708809 | Chr7 | 23243225 | |
| Translocation | Chr8 | 25082588 | Chr4 | 19215213 | |
| Translocation | Chr12 | 27052698 | Chr4 | 11544347 | LOC_Os12g43590 |
