Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2467 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2467 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 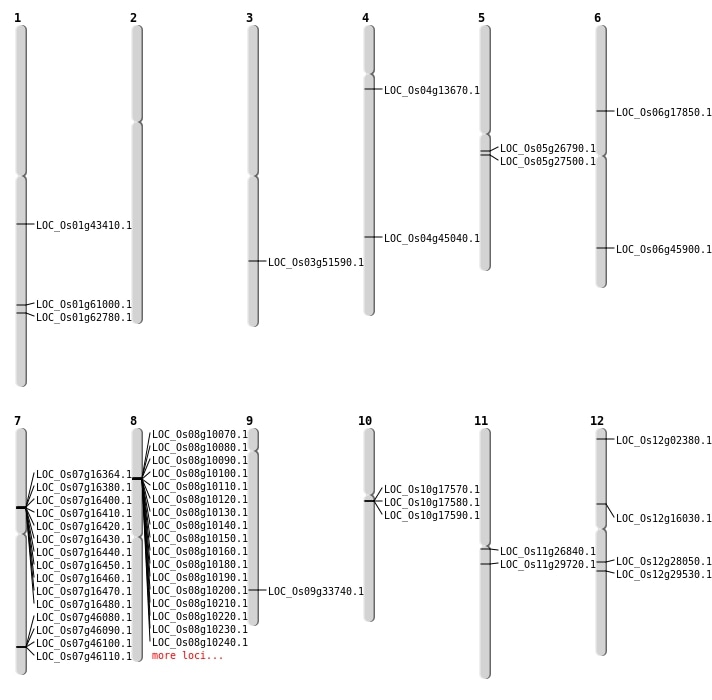
|
| Variant Information |
|---|
Single base substitutions: 47
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 15684632 | C-A | HET | ||
| SBS | Chr1 | 16304884 | C-T | HET | ||
| SBS | Chr1 | 17610311 | A-G | HET | ||
| SBS | Chr1 | 20020246 | C-T | HET | ||
| SBS | Chr1 | 24841153 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g43410 |
| SBS | Chr1 | 36357400 | A-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os01g62780 |
| SBS | Chr1 | 39296789 | C-T | HET | ||
| SBS | Chr10 | 21790708 | C-A | Homo | ||
| SBS | Chr11 | 10882350 | G-A | Homo | ||
| SBS | Chr11 | 11133786 | A-G | HET | ||
| SBS | Chr11 | 26724172 | C-T | HET | ||
| SBS | Chr11 | 26872435 | G-A | HET | ||
| SBS | Chr11 | 5267825 | T-A | HET | ||
| SBS | Chr12 | 16544003 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g28050 |
| SBS | Chr12 | 17595026 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g29530 |
| SBS | Chr12 | 25038381 | A-C | Homo | ||
| SBS | Chr12 | 5399582 | C-T | Homo | ||
| SBS | Chr12 | 8361672 | G-C | HET | ||
| SBS | Chr2 | 10417295 | T-C | HET | ||
| SBS | Chr2 | 25526354 | C-T | Homo | ||
| SBS | Chr2 | 33245249 | G-T | HET | ||
| SBS | Chr2 | 33669720 | T-C | HET | ||
| SBS | Chr2 | 33677822 | A-C | HET | ||
| SBS | Chr2 | 9192628 | C-G | HET | ||
| SBS | Chr3 | 20986730 | A-G | HET | ||
| SBS | Chr3 | 27326751 | G-T | HET | ||
| SBS | Chr3 | 5356372 | A-C | HET | ||
| SBS | Chr4 | 203647 | T-C | Homo | ||
| SBS | Chr4 | 35325679 | A-T | HET | ||
| SBS | Chr4 | 7630870 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g13670 |
| SBS | Chr5 | 10730724 | T-C | HET | ||
| SBS | Chr5 | 11781102 | T-C | HET | ||
| SBS | Chr5 | 15539047 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os05g26790 |
| SBS | Chr5 | 15996237 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os05g27500 |
| SBS | Chr5 | 1721619 | G-C | HET | ||
| SBS | Chr5 | 26124415 | C-T | HET | ||
| SBS | Chr6 | 15195091 | A-T | HET | ||
| SBS | Chr6 | 18080061 | C-T | HET | ||
| SBS | Chr6 | 26952880 | C-T | HET | ||
| SBS | Chr7 | 11438174 | A-C | HET | ||
| SBS | Chr7 | 14397053 | A-T | HET | ||
| SBS | Chr7 | 410103 | G-A | HET | ||
| SBS | Chr8 | 11647988 | G-T | HET | ||
| SBS | Chr8 | 5820597 | C-T | HET | ||
| SBS | Chr8 | 7672274 | C-T | HET | ||
| SBS | Chr9 | 14643561 | C-T | HET | ||
| SBS | Chr9 | 9696165 | C-T | Homo |
Deletions: 13
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr12 | 779168 | 779170 | 2 | LOC_Os12g02380 |
| Deletion | Chr10 | 1733907 | 1733908 | 1 | |
| Deletion | Chr8 | 5841001 | 5947000 | 105999 | 18 |
| Deletion | Chr10 | 8862001 | 8885000 | 22999 | 3 |
| Deletion | Chr11 | 9122976 | 9122980 | 4 | |
| Deletion | Chr7 | 9562001 | 9657000 | 94999 | 11 |
| Deletion | Chr5 | 11703547 | 11703550 | 3 | |
| Deletion | Chr11 | 17236914 | 17236937 | 23 | LOC_Os11g29720 |
| Deletion | Chr7 | 23431543 | 23431544 | 1 | |
| Deletion | Chr7 | 27501001 | 27523000 | 21999 | 4 |
| Deletion | Chr3 | 29521709 | 29521751 | 42 | LOC_Os03g51590 |
| Deletion | Chr1 | 35294308 | 35294315 | 7 | LOC_Os01g61000 |
| Deletion | Chr1 | 35729551 | 35729553 | 2 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 22279497 | 22279498 | 2 | |
| Insertion | Chr10 | 20923822 | 20923822 | 1 | |
| Insertion | Chr6 | 9547274 | 9547279 | 6 |
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr1 | 9780259 | 10042698 | |
| Inversion | Chr4 | 15111406 | 15974938 | |
| Inversion | Chr6 | 27161697 | 27787146 | 2 |
Translocations: 7
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 1589518 | Chr2 | 9833429 | |
| Translocation | Chr12 | 9135216 | Chr6 | 10364027 | 2 |
| Translocation | Chr9 | 9357599 | Chr6 | 29946116 | 2 |
| Translocation | Chr4 | 15111317 | Chr1 | 9780252 | |
| Translocation | Chr11 | 15418912 | Chr9 | 16554883 | LOC_Os11g26840 |
| Translocation | Chr11 | 15421322 | Chr9 | 16554887 | LOC_Os11g26850 |
| Translocation | Chr9 | 19929083 | Chr4 | 26648533 | 2 |
