Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2477 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2477 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 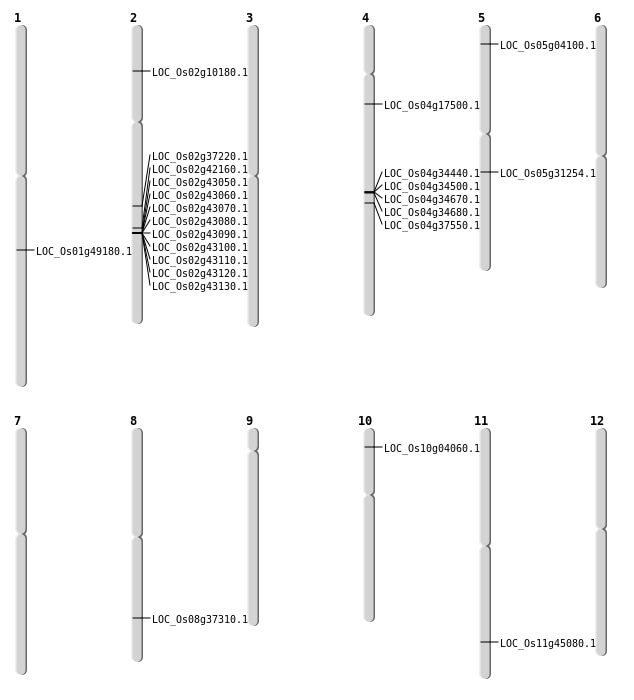
|
| Variant Information |
|---|
Single base substitutions: 44
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 26623929 | T-G | HET | ||
| SBS | Chr1 | 35843641 | C-T | HET | ||
| SBS | Chr1 | 37326908 | T-C | HET | ||
| SBS | Chr10 | 11861501 | C-A | HET | ||
| SBS | Chr10 | 12407415 | C-T | Homo | ||
| SBS | Chr10 | 3014714 | G-A | Homo | ||
| SBS | Chr10 | 7060773 | C-G | HET | ||
| SBS | Chr11 | 19661570 | C-G | HET | ||
| SBS | Chr11 | 23017890 | T-G | HET | ||
| SBS | Chr11 | 26837501 | T-C | HET | ||
| SBS | Chr11 | 27279845 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os11g45080 |
| SBS | Chr11 | 2886537 | C-T | HET | ||
| SBS | Chr12 | 10103671 | C-A | HET | ||
| SBS | Chr12 | 1541662 | C-T | HET | ||
| SBS | Chr12 | 17000924 | G-T | HET | ||
| SBS | Chr12 | 20244983 | G-A | HET | ||
| SBS | Chr12 | 6610514 | C-T | HET | ||
| SBS | Chr2 | 10396762 | A-C | HET | ||
| SBS | Chr2 | 10403395 | C-A | HET | ||
| SBS | Chr2 | 1333481 | T-A | Homo | ||
| SBS | Chr2 | 25359756 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g42160 |
| SBS | Chr2 | 7270807 | C-T | HET | ||
| SBS | Chr3 | 23033821 | C-T | Homo | ||
| SBS | Chr3 | 34696873 | G-A | HET | ||
| SBS | Chr3 | 4931462 | G-A | Homo | ||
| SBS | Chr3 | 6138313 | T-A | Homo | ||
| SBS | Chr3 | 7306957 | G-T | Homo | ||
| SBS | Chr4 | 20851775 | T-C | HET | ||
| SBS | Chr4 | 22348189 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g37550 |
| SBS | Chr4 | 22348191 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os04g37550 |
| SBS | Chr4 | 31627700 | C-A | Homo | ||
| SBS | Chr4 | 33047136 | G-T | Homo | ||
| SBS | Chr4 | 9575987 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os04g17500 |
| SBS | Chr5 | 23366997 | G-A | HET | ||
| SBS | Chr6 | 14372356 | C-T | HET | ||
| SBS | Chr6 | 25724058 | G-A | Homo | ||
| SBS | Chr6 | 27697596 | G-A | Homo | ||
| SBS | Chr6 | 4134614 | G-A | HET | ||
| SBS | Chr6 | 6658481 | G-A | HET | ||
| SBS | Chr7 | 23012483 | G-A | Homo | ||
| SBS | Chr7 | 25976380 | G-C | HET | ||
| SBS | Chr8 | 23578833 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os08g37310 |
| SBS | Chr8 | 9302987 | C-T | Homo | ||
| SBS | Chr9 | 9743375 | G-T | HET |
Deletions: 19
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr5 | 1843424 | 1843436 | 12 | LOC_Os05g04100 |
| Deletion | Chr10 | 1874867 | 1874873 | 6 | LOC_Os10g04060 |
| Deletion | Chr2 | 4123472 | 4123476 | 4 | |
| Deletion | Chr3 | 4674438 | 4674439 | 1 | |
| Deletion | Chr2 | 5332174 | 5332183 | 9 | LOC_Os02g10180 |
| Deletion | Chr6 | 7813942 | 7813945 | 3 | |
| Deletion | Chr4 | 9557770 | 9557778 | 8 | |
| Deletion | Chr9 | 9941368 | 9941369 | 1 | |
| Deletion | Chr11 | 13931333 | 13931335 | 2 | |
| Deletion | Chr12 | 17907922 | 17907928 | 6 | |
| Deletion | Chr7 | 18266897 | 18266898 | 1 | |
| Deletion | Chr4 | 19517444 | 19517452 | 8 | |
| Deletion | Chr4 | 20967001 | 20987000 | 19999 | 2 |
| Deletion | Chr12 | 21546165 | 21546168 | 3 | |
| Deletion | Chr2 | 22489134 | 22489135 | 1 | LOC_Os02g37220 |
| Deletion | Chr2 | 25920001 | 25978000 | 57999 | 9 |
| Deletion | Chr5 | 28784384 | 28784392 | 8 | |
| Deletion | Chr3 | 33758570 | 33758577 | 7 | |
| Deletion | Chr3 | 34560950 | 34560958 | 8 |
Insertions: 5
Inversions: 8
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr12 | 4251145 | 4669838 | |
| Inversion | Chr12 | 4251151 | 4669841 | |
| Inversion | Chr5 | 18171743 | 18606149 | LOC_Os05g31254 |
| Inversion | Chr5 | 18171750 | 18606155 | LOC_Os05g31254 |
| Inversion | Chr4 | 20853895 | 20888982 | 2 |
| Inversion | Chr4 | 20853897 | 20888993 | 2 |
| Inversion | Chr1 | 28268318 | 28994682 | LOC_Os01g49180 |
| Inversion | Chr1 | 28276045 | 28994685 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr10 | 18963098 | Chr1 | 27209441 | |
| Translocation | Chr2 | 25284977 | Chr1 | 12528665 |
