Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2479 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2479 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 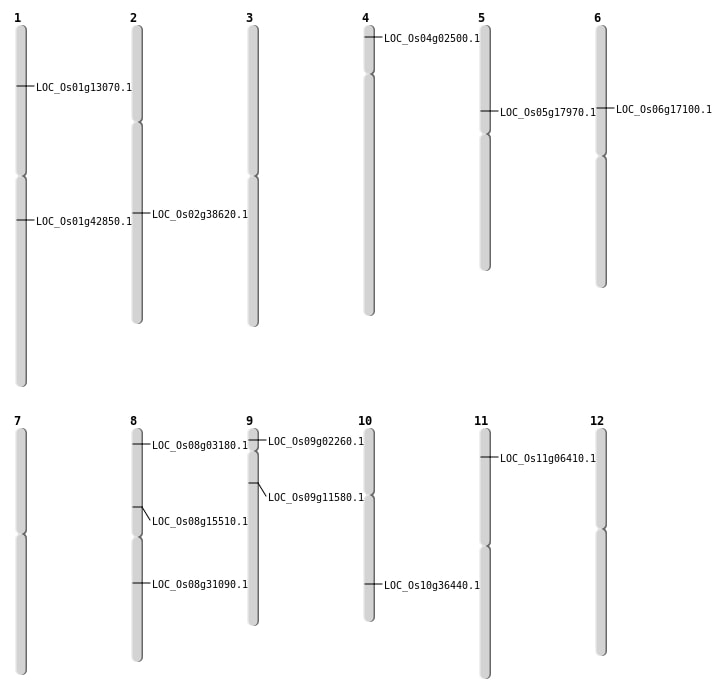
|
| Variant Information |
|---|
Single base substitutions: 47
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 24829076 | C-T | HET | ||
| SBS | Chr1 | 24829079 | T-A | HET | ||
| SBS | Chr1 | 24829082 | G-A | HET | ||
| SBS | Chr1 | 34011789 | G-A | HET | ||
| SBS | Chr1 | 4899764 | A-T | HET | ||
| SBS | Chr1 | 608843 | T-C | HET | ||
| SBS | Chr10 | 19497644 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os10g36440 |
| SBS | Chr11 | 18079249 | T-A | HET | ||
| SBS | Chr11 | 3093856 | T-G | HET | NON_SYNONYMOUS_CODING | LOC_Os11g06410 |
| SBS | Chr11 | 774148 | A-G | Homo | ||
| SBS | Chr12 | 12662798 | G-A | Homo | ||
| SBS | Chr12 | 19633101 | C-T | Homo | ||
| SBS | Chr12 | 27305253 | T-C | HET | ||
| SBS | Chr12 | 9707158 | G-A | HET | ||
| SBS | Chr2 | 17058106 | G-A | HET | ||
| SBS | Chr2 | 21027792 | A-G | HET | ||
| SBS | Chr2 | 22011481 | C-T | HET | ||
| SBS | Chr2 | 23343033 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os02g38620 |
| SBS | Chr2 | 75568 | T-C | Homo | ||
| SBS | Chr3 | 23699864 | C-T | HET | ||
| SBS | Chr3 | 31621087 | T-C | HET | ||
| SBS | Chr3 | 4923644 | G-A | HET | ||
| SBS | Chr3 | 6288325 | C-A | HET | ||
| SBS | Chr3 | 6288329 | G-A | HET | ||
| SBS | Chr3 | 6288330 | C-A | HET | ||
| SBS | Chr3 | 7035662 | G-A | HET | ||
| SBS | Chr3 | 9238544 | T-C | HET | ||
| SBS | Chr4 | 10564072 | G-A | HET | ||
| SBS | Chr4 | 15368376 | A-G | HET | ||
| SBS | Chr4 | 18426121 | C-T | HET | ||
| SBS | Chr4 | 26718284 | G-A | HET | ||
| SBS | Chr4 | 5268243 | A-T | HET | ||
| SBS | Chr4 | 8826249 | C-A | HET | ||
| SBS | Chr5 | 10329747 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os05g17970 |
| SBS | Chr5 | 9928927 | A-G | HET | ||
| SBS | Chr6 | 2838128 | A-G | HET | ||
| SBS | Chr6 | 4861483 | G-A | HET | ||
| SBS | Chr7 | 17732185 | C-T | HET | ||
| SBS | Chr7 | 20499252 | G-A | Homo | ||
| SBS | Chr8 | 1127362 | G-C | HET | ||
| SBS | Chr8 | 1801665 | T-C | HET | ||
| SBS | Chr8 | 19207274 | A-C | HET | NON_SYNONYMOUS_CODING | LOC_Os08g31090 |
| SBS | Chr8 | 25751840 | C-A | HET | ||
| SBS | Chr9 | 13579940 | G-C | HET | ||
| SBS | Chr9 | 14374182 | C-A | HET | ||
| SBS | Chr9 | 6459630 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os09g11580 |
| SBS | Chr9 | 916386 | T-G | HET | NON_SYNONYMOUS_CODING | LOC_Os09g02260 |
Deletions: 17
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr4 | 906690 | 906692 | 2 | LOC_Os04g02500 |
| Deletion | Chr2 | 1120980 | 1120981 | 1 | |
| Deletion | Chr2 | 4536027 | 4536028 | 1 | |
| Deletion | Chr4 | 4899846 | 4899853 | 7 | |
| Deletion | Chr1 | 7276506 | 7276508 | 2 | LOC_Os01g13070 |
| Deletion | Chr12 | 7845340 | 7845356 | 16 | |
| Deletion | Chr9 | 9139061 | 9139071 | 10 | |
| Deletion | Chr8 | 9434926 | 9434928 | 2 | LOC_Os08g15510 |
| Deletion | Chr6 | 9913567 | 9913574 | 7 | LOC_Os06g17100 |
| Deletion | Chr4 | 9975325 | 9975326 | 1 | |
| Deletion | Chr6 | 10704890 | 10704900 | 10 | |
| Deletion | Chr8 | 17115670 | 17115682 | 12 | |
| Deletion | Chr5 | 17184534 | 17184540 | 6 | |
| Deletion | Chr2 | 22006453 | 22006464 | 11 | |
| Deletion | Chr4 | 26310286 | 26310301 | 15 | |
| Deletion | Chr7 | 28463304 | 28463305 | 1 | |
| Deletion | Chr5 | 28778124 | 28778127 | 3 |
Insertions: 6
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr1 | 24383674 | 24660388 | LOC_Os01g42850 |
| Inversion | Chr1 | 24383677 | 24447883 | LOC_Os01g42850 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 917459 | Chr8 | 1472857 | 2 |
| Translocation | Chr9 | 917879 | Chr8 | 1472858 | 2 |
| Translocation | Chr11 | 23368411 | Chr5 | 13251370 |
