Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2498 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2498 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 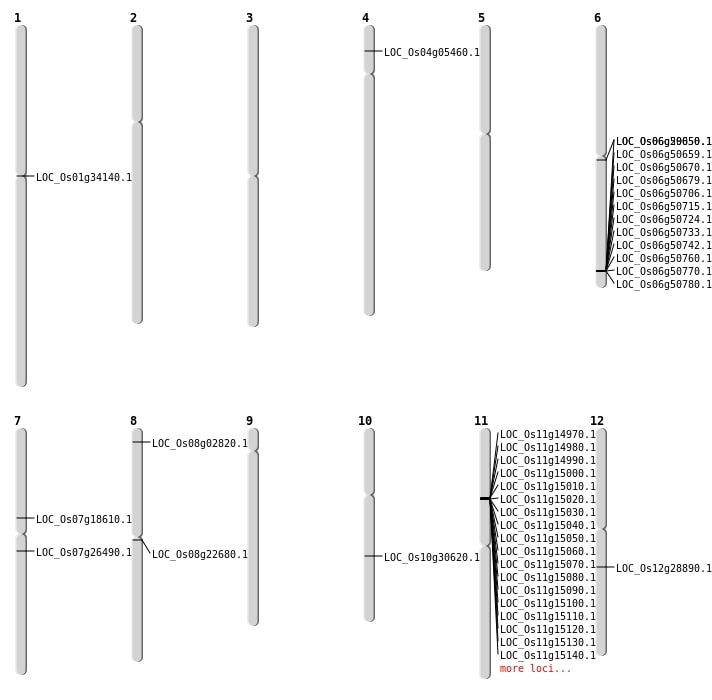
|
| Variant Information |
|---|
Single base substitutions: 43
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 10077784 | A-T | HET | ||
| SBS | Chr1 | 16998445 | T-C | HET | ||
| SBS | Chr1 | 17090789 | G-A | Homo | ||
| SBS | Chr1 | 17090790 | G-A | Homo | ||
| SBS | Chr1 | 35041848 | C-T | HET | ||
| SBS | Chr1 | 36967912 | T-G | HET | ||
| SBS | Chr1 | 7710828 | C-T | HET | ||
| SBS | Chr10 | 15941827 | T-G | HET | NON_SYNONYMOUS_CODING | LOC_Os10g30620 |
| SBS | Chr10 | 15982657 | C-A | HET | ||
| SBS | Chr10 | 21786126 | A-G | HET | ||
| SBS | Chr11 | 15179970 | C-T | HET | ||
| SBS | Chr11 | 19559078 | A-T | HET | ||
| SBS | Chr11 | 25637723 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os11g42590 |
| SBS | Chr11 | 3356001 | A-T | HET | ||
| SBS | Chr12 | 10317222 | T-C | HET | ||
| SBS | Chr12 | 17082557 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g28890 |
| SBS | Chr12 | 2478991 | T-A | Homo | ||
| SBS | Chr12 | 2666454 | G-A | Homo | ||
| SBS | Chr12 | 4325257 | C-A | HET | ||
| SBS | Chr12 | 5505293 | G-C | HET | ||
| SBS | Chr12 | 7730374 | T-C | HET | ||
| SBS | Chr2 | 20114884 | C-T | HET | ||
| SBS | Chr2 | 20902230 | T-A | HET | ||
| SBS | Chr2 | 23827789 | C-T | HET | ||
| SBS | Chr2 | 5980765 | G-T | HET | ||
| SBS | Chr3 | 352718 | G-T | HET | ||
| SBS | Chr4 | 22569268 | A-T | HET | ||
| SBS | Chr4 | 4379698 | G-A | HET | ||
| SBS | Chr5 | 14147920 | G-A | HET | ||
| SBS | Chr5 | 16360028 | G-T | HET | ||
| SBS | Chr5 | 17264713 | T-C | HET | ||
| SBS | Chr5 | 9470733 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g16670 |
| SBS | Chr6 | 15914095 | G-T | HET | ||
| SBS | Chr7 | 10998431 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g18610 |
| SBS | Chr7 | 15238251 | A-G | Homo | NON_SYNONYMOUS_CODING | LOC_Os07g26490 |
| SBS | Chr7 | 24068638 | C-G | HET | ||
| SBS | Chr8 | 10945899 | G-A | HET | ||
| SBS | Chr8 | 1183354 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os08g02820 |
| SBS | Chr8 | 8807498 | C-T | Homo | ||
| SBS | Chr9 | 11377348 | C-T | Homo | ||
| SBS | Chr9 | 16551719 | A-T | HET | ||
| SBS | Chr9 | 17964797 | T-A | HET | ||
| SBS | Chr9 | 9348074 | C-T | HET |
Deletions: 9
Insertions: 5
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr12 | 3959845 | 3959846 | 2 | |
| Insertion | Chr2 | 32801646 | 32801646 | 1 | |
| Insertion | Chr6 | 7629182 | 7629183 | 2 | |
| Insertion | Chr8 | 13642396 | 13642396 | 1 | LOC_Os08g22680 |
| Insertion | Chr9 | 5809357 | 5809358 | 2 |
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr1 | 18807135 | 18841789 | LOC_Os01g34140 |
Translocations: 4
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 4920900 | Chr1 | 19588631 | |
| Translocation | Chr6 | 16552093 | Chr4 | 2748318 | 2 |
| Translocation | Chr8 | 19774183 | Chr5 | 12662789 | |
| Translocation | Chr12 | 27043817 | Chr11 | 20669201 |
