Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2530-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2530-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 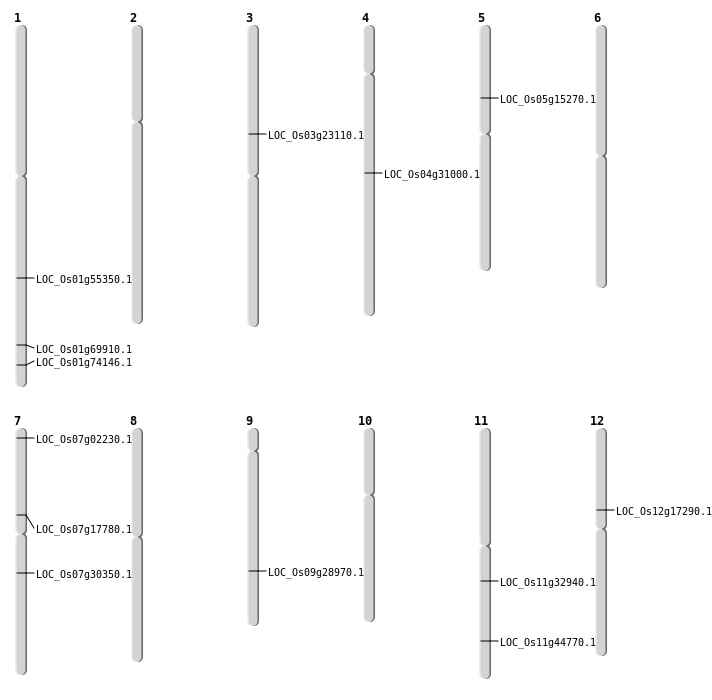
|
| Variant Information |
|---|
Single base substitutions: 67
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 10001963 | C-T | Homo | ||
| SBS | Chr1 | 13806447 | T-A | HET | ||
| SBS | Chr1 | 19467871 | T-G | HET | ||
| SBS | Chr1 | 21185871 | T-C | HET | ||
| SBS | Chr1 | 24760932 | G-A | HET | ||
| SBS | Chr1 | 28764847 | G-A | HET | ||
| SBS | Chr1 | 29494400 | A-T | HET | ||
| SBS | Chr1 | 31862607 | C-T | HET | SPLICE_SITE_DONOR | LOC_Os01g55350 |
| SBS | Chr1 | 31862610 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os01g55350 |
| SBS | Chr1 | 32927602 | C-A | HET | ||
| SBS | Chr1 | 40400482 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g69910 |
| SBS | Chr1 | 42949979 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g74146 |
| SBS | Chr10 | 10282296 | C-T | HET | ||
| SBS | Chr10 | 1181874 | T-C | HET | ||
| SBS | Chr10 | 16679904 | G-A | Homo | ||
| SBS | Chr11 | 17241130 | C-A | HET | ||
| SBS | Chr11 | 2448756 | A-T | HET | ||
| SBS | Chr11 | 27094790 | C-T | HET | STOP_GAINED | LOC_Os11g44770 |
| SBS | Chr11 | 5323856 | T-C | HET | ||
| SBS | Chr12 | 10790095 | C-T | HET | ||
| SBS | Chr12 | 23794342 | T-A | HET | ||
| SBS | Chr12 | 24268854 | T-C | HET | ||
| SBS | Chr12 | 8504470 | G-A | HET | ||
| SBS | Chr12 | 8991030 | A-G | HET | ||
| SBS | Chr2 | 14016733 | C-A | HET | ||
| SBS | Chr2 | 14107638 | G-A | HET | ||
| SBS | Chr2 | 2667902 | A-C | Homo | ||
| SBS | Chr2 | 27793085 | G-A | HET | ||
| SBS | Chr2 | 32453312 | G-A | HET | ||
| SBS | Chr2 | 5538029 | T-C | Homo | ||
| SBS | Chr3 | 14834340 | G-T | HET | ||
| SBS | Chr3 | 2336111 | C-A | HET | ||
| SBS | Chr3 | 4110149 | A-C | HET | ||
| SBS | Chr3 | 8084689 | C-T | HET | ||
| SBS | Chr3 | 9909611 | T-C | HET | ||
| SBS | Chr4 | 10365844 | C-T | HET | ||
| SBS | Chr4 | 10901030 | G-A | HET | ||
| SBS | Chr4 | 18523035 | C-T | HET | STOP_GAINED | LOC_Os04g31000 |
| SBS | Chr4 | 19121208 | C-T | HET | ||
| SBS | Chr4 | 21774360 | C-T | HET | ||
| SBS | Chr4 | 24045504 | G-A | HET | ||
| SBS | Chr4 | 34290328 | T-A | HET | ||
| SBS | Chr4 | 4134352 | C-T | HET | ||
| SBS | Chr4 | 5498987 | T-C | HET | ||
| SBS | Chr4 | 8008659 | G-T | HET | ||
| SBS | Chr5 | 12346882 | C-T | HET | ||
| SBS | Chr5 | 15198495 | A-T | HET | ||
| SBS | Chr5 | 20885730 | C-T | HET | ||
| SBS | Chr5 | 23098994 | T-C | HET | ||
| SBS | Chr5 | 3962042 | C-T | HET | ||
| SBS | Chr5 | 4285383 | G-A | HET | ||
| SBS | Chr6 | 19312814 | A-G | HET | ||
| SBS | Chr7 | 12331403 | G-A | HET | ||
| SBS | Chr7 | 13685678 | G-A | HET | ||
| SBS | Chr7 | 17963160 | G-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os07g30350 |
| SBS | Chr7 | 21242475 | A-G | HET | ||
| SBS | Chr7 | 2250178 | C-T | HET | ||
| SBS | Chr7 | 23145189 | C-A | HET | ||
| SBS | Chr7 | 23145190 | G-T | HET | ||
| SBS | Chr7 | 2764258 | G-C | HET | ||
| SBS | Chr7 | 707138 | A-G | Homo | NON_SYNONYMOUS_CODING | LOC_Os07g02230 |
| SBS | Chr8 | 11460556 | C-A | HET | ||
| SBS | Chr8 | 13646410 | T-A | HET | ||
| SBS | Chr8 | 16888952 | T-G | HET | ||
| SBS | Chr9 | 12816741 | C-A | Homo | ||
| SBS | Chr9 | 17619013 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os09g28970 |
| SBS | Chr9 | 4375595 | C-A | Homo |
Deletions: 13
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr7 | 711653 | 711664 | 11 | LOC_Os07g02230 |
| Deletion | Chr7 | 825875 | 825879 | 4 | |
| Deletion | Chr7 | 7492773 | 7492796 | 23 | |
| Deletion | Chr5 | 8659004 | 8659011 | 7 | LOC_Os05g15270 |
| Deletion | Chr12 | 9893643 | 9893648 | 5 | LOC_Os12g17290 |
| Deletion | Chr7 | 10506161 | 10506162 | 1 | LOC_Os07g17780 |
| Deletion | Chr8 | 11969940 | 11969949 | 9 | |
| Deletion | Chr3 | 13362876 | 13362877 | 1 | LOC_Os03g23110 |
| Deletion | Chr10 | 14374939 | 14374998 | 59 | |
| Deletion | Chr4 | 15155954 | 15155957 | 3 | |
| Deletion | Chr4 | 16400838 | 16400839 | 1 | |
| Deletion | Chr7 | 22514502 | 22514511 | 9 | |
| Deletion | Chr7 | 27955976 | 27955977 | 1 |
Insertions: 5
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr11 | 19453574 | 20359192 | LOC_Os11g32940 |
Translocations: 4
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 8941257 | Chr1 | 30237136 | |
| Translocation | Chr8 | 17137888 | Chr6 | 574442 | |
| Translocation | Chr11 | 17315139 | Chr7 | 8152570 | |
| Translocation | Chr10 | 19654441 | Chr4 | 6991375 |
