Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2532-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2532-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 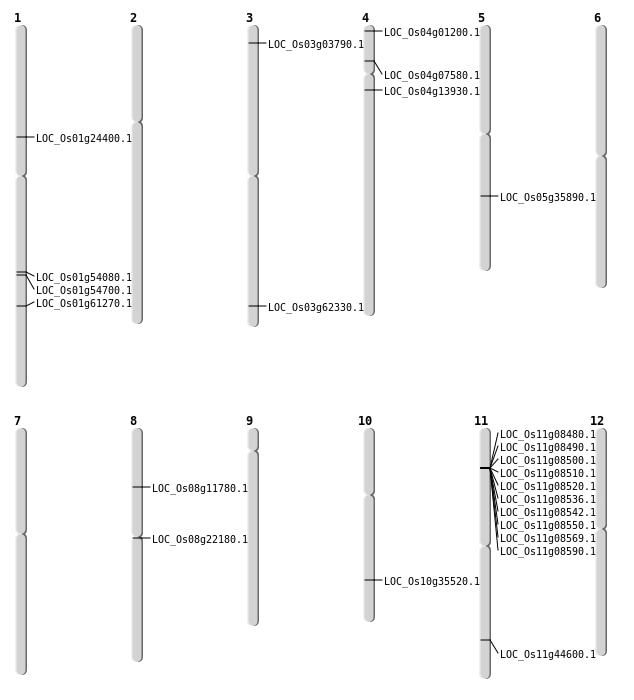
|
| Variant Information |
|---|
Single base substitutions: 56
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 13754678 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g24400 |
| SBS | Chr1 | 15250914 | G-A | Homo | ||
| SBS | Chr1 | 21175729 | T-G | Homo | ||
| SBS | Chr1 | 35673161 | T-C | HET | ||
| SBS | Chr1 | 4589000 | T-A | Homo | ||
| SBS | Chr10 | 16604199 | C-A | HET | ||
| SBS | Chr10 | 19000472 | A-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os10g35520 |
| SBS | Chr10 | 19685111 | A-G | Homo | ||
| SBS | Chr10 | 19906079 | G-A | Homo | ||
| SBS | Chr10 | 20353515 | G-A | HET | ||
| SBS | Chr10 | 4467612 | A-G | Homo | ||
| SBS | Chr10 | 6899542 | C-A | HET | ||
| SBS | Chr11 | 17605259 | C-A | HET | ||
| SBS | Chr11 | 718187 | A-G | HET | ||
| SBS | Chr11 | 9029660 | G-A | HET | ||
| SBS | Chr12 | 10256042 | A-G | HET | ||
| SBS | Chr12 | 17340270 | C-T | HET | ||
| SBS | Chr12 | 19769488 | C-T | HET | ||
| SBS | Chr12 | 20265836 | T-C | Homo | ||
| SBS | Chr12 | 2167369 | T-A | HET | ||
| SBS | Chr12 | 530904 | C-A | HET | ||
| SBS | Chr2 | 21094546 | T-C | HET | ||
| SBS | Chr2 | 24908678 | A-G | HET | ||
| SBS | Chr2 | 31073761 | G-T | HET | ||
| SBS | Chr2 | 31073762 | C-G | HET | ||
| SBS | Chr2 | 32047420 | G-A | HET | ||
| SBS | Chr3 | 12058563 | G-A | HET | ||
| SBS | Chr3 | 17373061 | C-T | HET | ||
| SBS | Chr3 | 1850871 | G-A | HET | ||
| SBS | Chr3 | 24671974 | G-T | HET | ||
| SBS | Chr3 | 29366049 | T-A | HET | ||
| SBS | Chr3 | 29419488 | C-A | HET | ||
| SBS | Chr3 | 35310576 | T-C | HET | ||
| SBS | Chr3 | 35310578 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os03g62330 |
| SBS | Chr3 | 7824293 | C-A | Homo | ||
| SBS | Chr4 | 12633186 | T-C | Homo | ||
| SBS | Chr4 | 25834734 | C-T | Homo | ||
| SBS | Chr4 | 30836904 | C-T | Homo | ||
| SBS | Chr4 | 4033350 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g07580 |
| SBS | Chr4 | 7781153 | G-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os04g13930 |
| SBS | Chr5 | 13021081 | C-T | HET | ||
| SBS | Chr5 | 23513317 | G-T | HET | ||
| SBS | Chr5 | 4948002 | T-C | HET | ||
| SBS | Chr6 | 6896498 | C-G | Homo | ||
| SBS | Chr7 | 11174262 | G-T | HET | ||
| SBS | Chr8 | 13381137 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g22180 |
| SBS | Chr8 | 13509310 | G-A | HET | ||
| SBS | Chr8 | 17481549 | A-T | HET | ||
| SBS | Chr8 | 21686661 | T-C | HET | ||
| SBS | Chr8 | 24278158 | C-G | HET | ||
| SBS | Chr8 | 6902174 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g11780 |
| SBS | Chr8 | 7441935 | A-G | HET | ||
| SBS | Chr9 | 11382828 | C-A | HET | ||
| SBS | Chr9 | 5354338 | G-A | HET | ||
| SBS | Chr9 | 6486653 | C-G | HET | ||
| SBS | Chr9 | 6529411 | G-A | HET |
Deletions: 14
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr4 | 144528 | 144532 | 4 | LOC_Os04g01200 |
| Deletion | Chr2 | 333476 | 333478 | 2 | |
| Deletion | Chr3 | 1707331 | 1707332 | 1 | LOC_Os03g03790 |
| Deletion | Chr6 | 4298285 | 4298287 | 2 | |
| Deletion | Chr11 | 4482001 | 4567000 | 84999 | 10 |
| Deletion | Chr12 | 4484327 | 4484338 | 11 | |
| Deletion | Chr11 | 6358279 | 6358282 | 3 | |
| Deletion | Chr1 | 7465223 | 7465224 | 1 | |
| Deletion | Chr6 | 13856786 | 13856788 | 2 | |
| Deletion | Chr10 | 14948665 | 14948666 | 1 | |
| Deletion | Chr3 | 16999129 | 16999131 | 2 | |
| Deletion | Chr8 | 20599442 | 20599447 | 5 | |
| Deletion | Chr5 | 21289060 | 21289061 | 1 | LOC_Os05g35890 |
| Deletion | Chr2 | 24990426 | 24990427 | 1 |
Insertions: 2
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr3 | 11892654 | 11892654 | 1 | |
| Insertion | Chr4 | 8953676 | 8953701 | 26 |
Inversions: 6
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr8 | 19754201 | Chr1 | 24031476 | |
| Translocation | Chr11 | 26972550 | Chr1 | 35452941 | 2 |
