Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2537-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2537-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 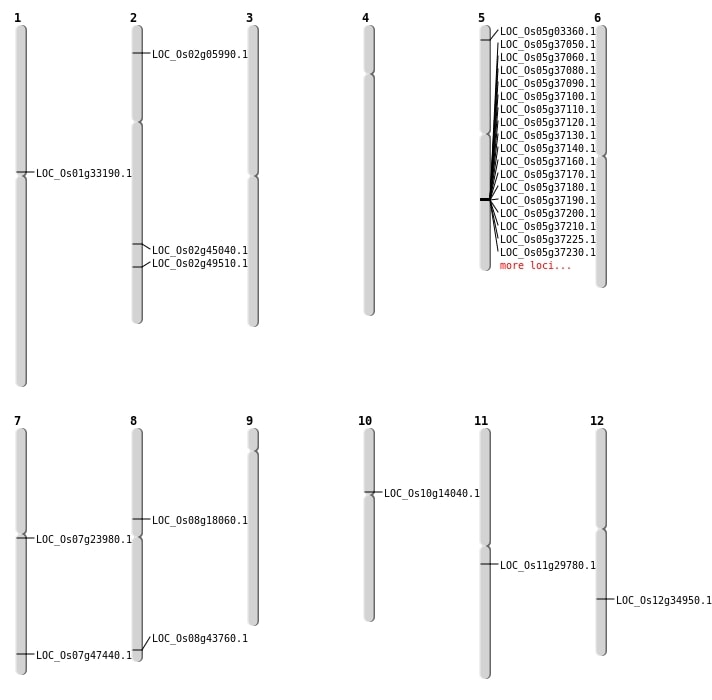
|
| Variant Information |
|---|
Single base substitutions: 45
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 15140998 | G-A | HET | ||
| SBS | Chr1 | 17051369 | C-A | Homo | ||
| SBS | Chr1 | 18273970 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g33190 |
| SBS | Chr1 | 28903348 | A-T | HET | ||
| SBS | Chr10 | 2734711 | G-C | HET | ||
| SBS | Chr10 | 7638371 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os10g14040 |
| SBS | Chr10 | 7800580 | C-T | HET | ||
| SBS | Chr10 | 7800584 | C-T | HET | ||
| SBS | Chr11 | 2390374 | G-A | HET | ||
| SBS | Chr11 | 292861 | A-C | HET | ||
| SBS | Chr12 | 11854064 | T-A | Homo | ||
| SBS | Chr12 | 14771771 | T-A | Homo | ||
| SBS | Chr2 | 19475055 | C-T | HET | ||
| SBS | Chr2 | 27298600 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g45040 |
| SBS | Chr2 | 29220120 | T-C | HET | ||
| SBS | Chr2 | 2989009 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g05990 |
| SBS | Chr2 | 7786712 | G-A | Homo | ||
| SBS | Chr3 | 30351838 | T-A | HET | ||
| SBS | Chr3 | 30718872 | C-T | HET | ||
| SBS | Chr3 | 5011799 | C-T | Homo | ||
| SBS | Chr4 | 14445601 | G-T | Homo | ||
| SBS | Chr4 | 18743989 | T-A | Homo | ||
| SBS | Chr4 | 35215590 | G-A | HET | ||
| SBS | Chr4 | 7993428 | G-A | HET | ||
| SBS | Chr5 | 12848238 | G-A | HET | ||
| SBS | Chr5 | 1378859 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os05g03360 |
| SBS | Chr5 | 6478416 | G-T | HET | ||
| SBS | Chr6 | 11963254 | G-C | HET | ||
| SBS | Chr6 | 21711130 | G-A | HET | ||
| SBS | Chr6 | 7891970 | A-G | HET | ||
| SBS | Chr6 | 8857031 | G-A | HET | ||
| SBS | Chr6 | 9692444 | C-T | HET | ||
| SBS | Chr7 | 10008092 | A-T | HET | ||
| SBS | Chr7 | 10552363 | T-A | Homo | ||
| SBS | Chr7 | 10694165 | C-A | Homo | ||
| SBS | Chr7 | 20394152 | G-A | HET | ||
| SBS | Chr7 | 27580146 | A-T | HET | ||
| SBS | Chr7 | 27580150 | T-G | HET | ||
| SBS | Chr7 | 3951846 | A-C | Homo | ||
| SBS | Chr8 | 11065142 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os08g18060 |
| SBS | Chr8 | 14513496 | C-T | HET | ||
| SBS | Chr8 | 27645332 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g43760 |
| SBS | Chr8 | 7655897 | C-T | HET | ||
| SBS | Chr9 | 12272587 | T-G | HET | ||
| SBS | Chr9 | 15002414 | G-T | Homo |
Deletions: 18
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr9 | 1551630 | 1551636 | 6 | |
| Deletion | Chr12 | 2120729 | 2120735 | 6 | |
| Deletion | Chr11 | 2633638 | 2633645 | 7 | |
| Deletion | Chr10 | 7255521 | 7255525 | 4 | |
| Deletion | Chr12 | 8978051 | 8978053 | 2 | |
| Deletion | Chr5 | 9149537 | 9149546 | 9 | |
| Deletion | Chr11 | 13631614 | 13631623 | 9 | |
| Deletion | Chr2 | 15063697 | 15063698 | 1 | |
| Deletion | Chr3 | 17716922 | 17716924 | 2 | |
| Deletion | Chr8 | 18146954 | 18146955 | 1 | |
| Deletion | Chr12 | 21273451 | 21273459 | 8 | LOC_Os12g34950 |
| Deletion | Chr5 | 21651001 | 21661000 | 9999 | 2 |
| Deletion | Chr5 | 21666001 | 21822000 | 155999 | 21 |
| Deletion | Chr5 | 21828001 | 22167000 | 338999 | 53 |
| Deletion | Chr6 | 23223845 | 23223848 | 3 | |
| Deletion | Chr2 | 25555096 | 25555226 | 130 | |
| Deletion | Chr7 | 28366635 | 28366642 | 7 | LOC_Os07g47440 |
| Deletion | Chr1 | 35915698 | 35915706 | 8 |
Insertions: 6
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr2 | 16137945 | 16137945 | 1 | |
| Insertion | Chr7 | 13576897 | 13576897 | 1 | LOC_Os07g23980 |
| Insertion | Chr8 | 11049434 | 11049434 | 1 | |
| Insertion | Chr9 | 12615143 | 12615143 | 1 | |
| Insertion | Chr9 | 2791278 | 2791278 | 1 | |
| Insertion | Chr9 | 3199189 | 3199195 | 7 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr11 | 17284194 | 17284409 | LOC_Os11g29780 |
| Inversion | Chr2 | 30175588 | 30259840 | 2 |
Translocations: 6
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr10 | 13446782 | Chr1 | 30260921 | 2 |
| Translocation | Chr10 | 13449173 | Chr1 | 30260916 | 2 |
| Translocation | Chr5 | 17627891 | Chr1 | 42980730 | |
| Translocation | Chr11 | 18033977 | Chr1 | 31269142 | |
| Translocation | Chr7 | 19416460 | Chr1 | 34821287 | |
| Translocation | Chr3 | 30824148 | Chr2 | 30175597 |
