Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2542-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2542-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 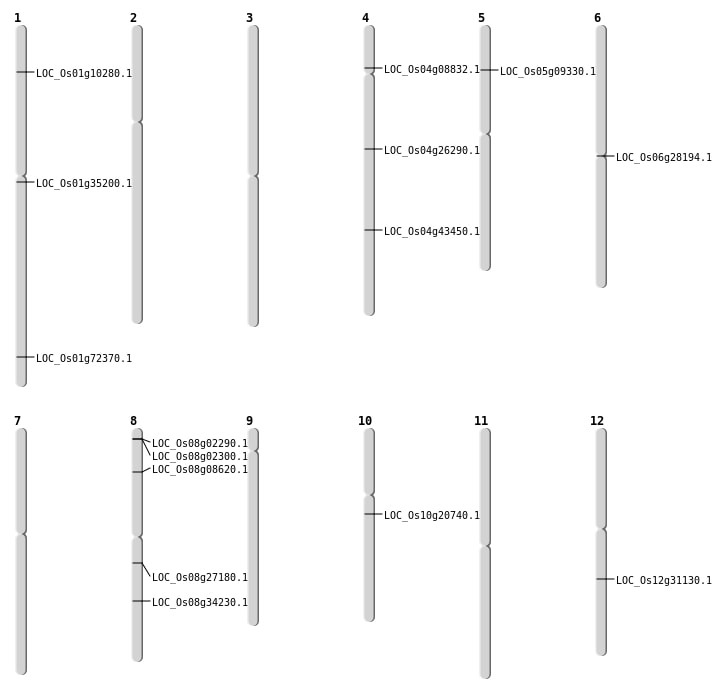
|
| Variant Information |
|---|
Single base substitutions: 49
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 13563200 | T-A | Homo | ||
| SBS | Chr1 | 20550288 | C-T | HET | ||
| SBS | Chr1 | 25661009 | A-G | HET | ||
| SBS | Chr1 | 27947256 | G-A | HET | ||
| SBS | Chr1 | 30046436 | T-A | HET | ||
| SBS | Chr1 | 39905634 | A-C | HET | ||
| SBS | Chr1 | 5411035 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g10280 |
| SBS | Chr10 | 10511969 | G-A | HET | STOP_GAINED | LOC_Os10g20740 |
| SBS | Chr10 | 12079542 | T-C | HET | ||
| SBS | Chr10 | 338164 | T-C | HET | ||
| SBS | Chr10 | 8160320 | T-G | HET | ||
| SBS | Chr11 | 12752488 | G-A | HET | ||
| SBS | Chr11 | 14318736 | T-C | HET | ||
| SBS | Chr11 | 16168869 | C-A | HET | ||
| SBS | Chr11 | 23883200 | G-A | HET | ||
| SBS | Chr12 | 15274547 | C-T | HET | ||
| SBS | Chr12 | 18738532 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os12g31130 |
| SBS | Chr12 | 19935426 | C-T | HET | ||
| SBS | Chr12 | 765820 | G-A | HET | ||
| SBS | Chr2 | 15831648 | C-G | HET | ||
| SBS | Chr2 | 31547664 | G-A | HET | ||
| SBS | Chr3 | 12038169 | A-G | HET | ||
| SBS | Chr3 | 12991373 | A-G | HET | ||
| SBS | Chr3 | 24751856 | A-G | HET | ||
| SBS | Chr3 | 33219764 | A-C | HET | ||
| SBS | Chr3 | 33415659 | C-T | HET | ||
| SBS | Chr3 | 6150959 | C-A | HET | ||
| SBS | Chr4 | 11138616 | C-T | HET | ||
| SBS | Chr4 | 15324557 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g26290 |
| SBS | Chr4 | 25714050 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g43450 |
| SBS | Chr4 | 4884547 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g08832 |
| SBS | Chr5 | 22259103 | G-A | HET | ||
| SBS | Chr6 | 12955659 | G-T | HET | ||
| SBS | Chr6 | 4045190 | C-G | HET | ||
| SBS | Chr7 | 10610761 | C-G | HET | ||
| SBS | Chr7 | 20630996 | C-A | HET | ||
| SBS | Chr7 | 4211655 | A-G | HET | ||
| SBS | Chr7 | 7834267 | T-G | |||
| SBS | Chr7 | 8498340 | G-A | HET | ||
| SBS | Chr7 | 9050216 | C-T | HET | ||
| SBS | Chr8 | 16597882 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g27180 |
| SBS | Chr8 | 21458443 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g34230 |
| SBS | Chr8 | 4984924 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g08620 |
| SBS | Chr9 | 11431652 | A-G | HET | ||
| SBS | Chr9 | 13007951 | G-A | Homo | ||
| SBS | Chr9 | 13613278 | C-A | HET | ||
| SBS | Chr9 | 16712149 | G-A | HET | ||
| SBS | Chr9 | 5705191 | T-C | Homo | ||
| SBS | Chr9 | 8791817 | G-A | HET |
Deletions: 13
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr8 | 826001 | 859000 | 32999 | 2 |
| Deletion | Chr7 | 2174000 | 2174004 | 4 | |
| Deletion | Chr2 | 2249822 | 2249823 | 1 | |
| Deletion | Chr12 | 2814262 | 2814263 | 1 | |
| Deletion | Chr2 | 2832702 | 2832709 | 7 | |
| Deletion | Chr1 | 9670186 | 9670191 | 5 | |
| Deletion | Chr4 | 10794076 | 10794077 | 1 | |
| Deletion | Chr2 | 23254980 | 23254981 | 1 | |
| Deletion | Chr4 | 23390785 | 23390807 | 22 | |
| Deletion | Chr7 | 28492110 | 28492111 | 1 | |
| Deletion | Chr4 | 28815940 | 28815944 | 4 | |
| Deletion | Chr2 | 30410403 | 30410412 | 9 | |
| Deletion | Chr1 | 41977822 | 41977824 | 2 | LOC_Os01g72370 |
Insertions: 7
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr2 | 2189220 | 2189823 | |
| Inversion | Chr6 | 16023351 | 16023579 | LOC_Os06g28194 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 15094758 | Chr1 | 12471727 | |
| Translocation | Chr6 | 23516057 | Chr1 | 19483726 | 2 |
| Translocation | Chr6 | 23516344 | Chr5 | 5212207 | 2 |
