Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2545-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2545-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 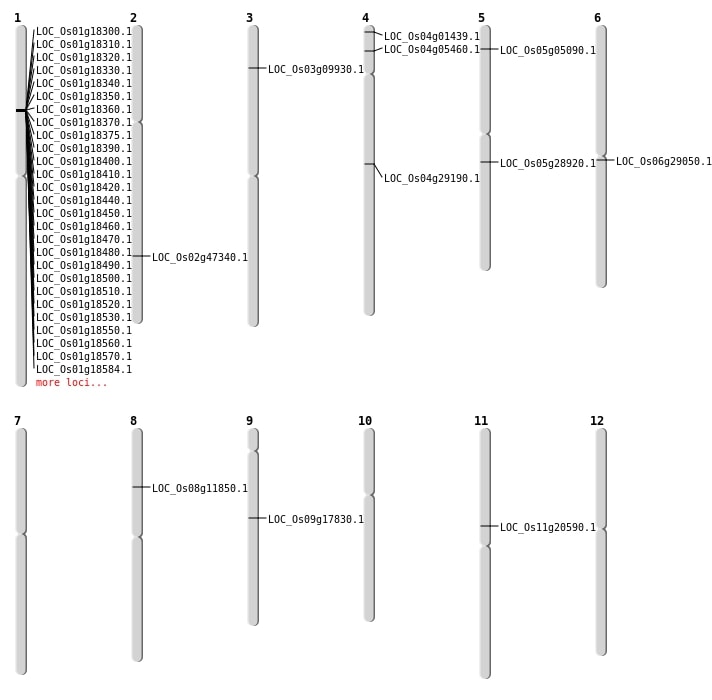
|
| Variant Information |
|---|
Single base substitutions: 54
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 15213687 | C-T | HET | ||
| SBS | Chr1 | 27977518 | G-A | HET | ||
| SBS | Chr1 | 30336445 | A-G | HET | ||
| SBS | Chr1 | 3905569 | T-A | Homo | ||
| SBS | Chr1 | 7831353 | A-C | HET | ||
| SBS | Chr11 | 11256251 | T-A | Homo | ||
| SBS | Chr11 | 11924804 | C-G | Homo | ||
| SBS | Chr11 | 16394235 | C-A | HET | ||
| SBS | Chr11 | 16394236 | A-G | HET | ||
| SBS | Chr11 | 2352135 | G-T | HET | ||
| SBS | Chr11 | 2612971 | G-A | HET | ||
| SBS | Chr11 | 8096130 | T-G | Homo | ||
| SBS | Chr11 | 8264479 | C-A | HET | ||
| SBS | Chr12 | 20180470 | G-T | HET | ||
| SBS | Chr2 | 11020083 | G-A | HET | ||
| SBS | Chr2 | 14894339 | A-G | HET | ||
| SBS | Chr2 | 17908175 | C-G | Homo | ||
| SBS | Chr2 | 28172743 | G-A | HET | ||
| SBS | Chr2 | 28904800 | G-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os02g47340 |
| SBS | Chr2 | 28983475 | T-C | HET | ||
| SBS | Chr2 | 31372390 | G-A | HET | ||
| SBS | Chr2 | 34874241 | T-A | Homo | ||
| SBS | Chr2 | 9951299 | A-G | Homo | ||
| SBS | Chr3 | 17641810 | A-C | HET | ||
| SBS | Chr3 | 24823818 | C-A | HET | ||
| SBS | Chr3 | 4947459 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g09930 |
| SBS | Chr4 | 10813959 | G-A | HET | ||
| SBS | Chr4 | 19351278 | C-T | HET | ||
| SBS | Chr4 | 20283628 | C-T | HET | ||
| SBS | Chr4 | 25807355 | G-A | HET | ||
| SBS | Chr4 | 25924254 | G-A | HET | ||
| SBS | Chr5 | 14609222 | G-A | HET | ||
| SBS | Chr5 | 16949862 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g28920 |
| SBS | Chr5 | 2474880 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g05090 |
| SBS | Chr5 | 2578163 | T-C | HET | ||
| SBS | Chr5 | 4260752 | G-A | HET | ||
| SBS | Chr5 | 4415813 | A-G | HET | ||
| SBS | Chr6 | 14799480 | G-A | HET | ||
| SBS | Chr6 | 15114655 | G-A | HET | ||
| SBS | Chr6 | 21239751 | G-A | HET | ||
| SBS | Chr6 | 23128326 | A-C | HET | ||
| SBS | Chr6 | 23784885 | T-A | HET | ||
| SBS | Chr6 | 8733118 | C-T | HET | ||
| SBS | Chr7 | 13860928 | C-T | HET | ||
| SBS | Chr7 | 19975824 | G-A | HET | ||
| SBS | Chr7 | 6831651 | C-T | Homo | ||
| SBS | Chr8 | 1021855 | A-C | HET | ||
| SBS | Chr8 | 13565191 | T-C | HET | ||
| SBS | Chr8 | 14714229 | G-A | HET | ||
| SBS | Chr8 | 15728206 | C-G | HET | ||
| SBS | Chr8 | 24070961 | A-C | HET | ||
| SBS | Chr8 | 9182140 | A-T | HET | ||
| SBS | Chr9 | 10907822 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os09g17830 |
| SBS | Chr9 | 11368143 | G-A | HET |
Deletions: 20
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr10 | 2286962 | 2286966 | 4 | |
| Deletion | Chr9 | 4630570 | 4630694 | 124 | |
| Deletion | Chr6 | 6575000 | 6575002 | 2 | |
| Deletion | Chr8 | 6871985 | 6871994 | 9 | |
| Deletion | Chr8 | 6952512 | 6952513 | 1 | LOC_Os08g11850 |
| Deletion | Chr1 | 10271001 | 10390000 | 118999 | 18 |
| Deletion | Chr1 | 10394001 | 10500000 | 105999 | 14 |
| Deletion | Chr11 | 11936301 | 11936302 | 1 | LOC_Os11g20590 |
| Deletion | Chr9 | 13098981 | 13098982 | 1 | |
| Deletion | Chr6 | 15284637 | 15284638 | 1 | |
| Deletion | Chr4 | 17306708 | 17306726 | 18 | LOC_Os04g29190 |
| Deletion | Chr12 | 17778566 | 17778569 | 3 | |
| Deletion | Chr10 | 21782853 | 21782854 | 1 | |
| Deletion | Chr7 | 23311732 | 23311733 | 1 | |
| Deletion | Chr6 | 25234429 | 25234433 | 4 | |
| Deletion | Chr8 | 26704437 | 26704439 | 2 | |
| Deletion | Chr8 | 27145230 | 27145245 | 15 | |
| Deletion | Chr8 | 27376485 | 27376487 | 2 | |
| Deletion | Chr4 | 27963533 | 27963546 | 13 | |
| Deletion | Chr1 | 31204205 | 31204209 | 4 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 5150421 | 5150422 | 2 | |
| Insertion | Chr10 | 12723232 | 12723232 | 1 | |
| Insertion | Chr11 | 22131445 | 22131449 | 5 | |
| Insertion | Chr8 | 95845 | 95856 | 12 |
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr4 | 308763 | 1087426 | LOC_Os04g01439 |
| Inversion | Chr3 | 18813810 | 19163155 | |
| Inversion | Chr3 | 18813826 | 19163160 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr6 | 16552093 | Chr4 | 2748318 | 2 |
| Translocation | Chr8 | 18006403 | Chr1 | 9104997 |
