Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2554-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2554-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 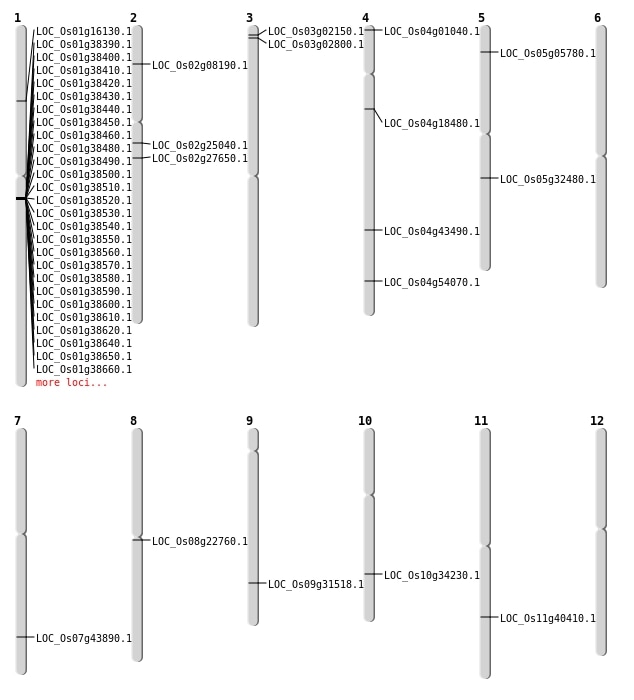
|
| Variant Information |
|---|
Single base substitutions: 64
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 11608995 | G-A | HET | ||
| SBS | Chr1 | 13993974 | C-G | HET | ||
| SBS | Chr1 | 28187548 | G-T | HET | ||
| SBS | Chr1 | 32461298 | A-T | HET | ||
| SBS | Chr1 | 35512069 | G-A | HET | ||
| SBS | Chr1 | 8012023 | A-G | HET | ||
| SBS | Chr10 | 11098367 | C-T | HET | ||
| SBS | Chr10 | 11261339 | T-A | HET | ||
| SBS | Chr11 | 21447951 | A-G | HET | ||
| SBS | Chr11 | 2945540 | C-T | HET | ||
| SBS | Chr12 | 4286552 | C-G | HET | ||
| SBS | Chr12 | 57679 | T-C | HET | ||
| SBS | Chr2 | 13427379 | G-T | HET | ||
| SBS | Chr2 | 14519132 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g25040 |
| SBS | Chr2 | 16382407 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g27650 |
| SBS | Chr2 | 18885343 | G-A | HET | ||
| SBS | Chr2 | 23248002 | G-T | HET | ||
| SBS | Chr2 | 27566678 | A-G | HET | ||
| SBS | Chr2 | 32225367 | G-T | Homo | ||
| SBS | Chr2 | 4337689 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os02g08190 |
| SBS | Chr2 | 4337693 | G-T | HET | ||
| SBS | Chr3 | 10187657 | C-T | HET | ||
| SBS | Chr3 | 10961508 | T-C | HET | ||
| SBS | Chr3 | 12276555 | T-A | HET | ||
| SBS | Chr3 | 13809606 | T-G | HET | ||
| SBS | Chr3 | 17280435 | C-T | HET | ||
| SBS | Chr3 | 21751371 | G-A | HET | ||
| SBS | Chr3 | 23033524 | C-T | HET | ||
| SBS | Chr3 | 28419030 | C-T | HET | ||
| SBS | Chr3 | 29401805 | C-A | HET | ||
| SBS | Chr3 | 31852753 | T-A | HET | ||
| SBS | Chr3 | 8922201 | G-T | HET | ||
| SBS | Chr3 | 8922202 | C-A | HET | ||
| SBS | Chr4 | 1289574 | G-A | HET | ||
| SBS | Chr4 | 13839328 | G-T | HET | ||
| SBS | Chr4 | 14875728 | T-A | HET | ||
| SBS | Chr4 | 14875730 | G-A | HET | ||
| SBS | Chr4 | 27651266 | T-C | HET | ||
| SBS | Chr4 | 3004756 | T-C | Homo | ||
| SBS | Chr4 | 31894381 | A-C | HET | ||
| SBS | Chr4 | 31894382 | T-A | HET | ||
| SBS | Chr5 | 13452254 | C-T | HET | ||
| SBS | Chr5 | 18582137 | A-G | HET | ||
| SBS | Chr5 | 18970562 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os05g32480 |
| SBS | Chr5 | 4058702 | C-T | HET | ||
| SBS | Chr6 | 26446239 | A-T | HET | ||
| SBS | Chr6 | 28004338 | G-A | HET | ||
| SBS | Chr6 | 3101326 | C-T | HET | ||
| SBS | Chr7 | 10578576 | C-A | HET | ||
| SBS | Chr7 | 19738042 | G-A | Homo | ||
| SBS | Chr7 | 2327212 | C-T | Homo | ||
| SBS | Chr7 | 2765420 | G-C | HET | ||
| SBS | Chr7 | 2860053 | G-A | Homo | ||
| SBS | Chr7 | 4836415 | G-A | Homo | ||
| SBS | Chr7 | 6690897 | T-C | HET | ||
| SBS | Chr7 | 7151559 | T-C | HET | ||
| SBS | Chr7 | 9664534 | C-A | HET | ||
| SBS | Chr8 | 16295004 | G-A | HET | ||
| SBS | Chr8 | 16741015 | G-A | HET | ||
| SBS | Chr8 | 17254227 | A-T | Homo | ||
| SBS | Chr8 | 5861407 | G-C | Homo | ||
| SBS | Chr9 | 12397787 | G-A | HET | ||
| SBS | Chr9 | 19031288 | T-G | Homo | NON_SYNONYMOUS_CODING | LOC_Os09g31518 |
| SBS | Chr9 | 19031289 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os09g31518 |
Deletions: 17
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr4 | 32873 | 32889 | 16 | LOC_Os04g01040 |
| Deletion | Chr8 | 1457519 | 1457533 | 14 | |
| Deletion | Chr10 | 2174975 | 2174979 | 4 | |
| Deletion | Chr9 | 6832418 | 6832424 | 6 | |
| Deletion | Chr12 | 7584391 | 7584402 | 11 | |
| Deletion | Chr11 | 9713142 | 9713153 | 11 | |
| Deletion | Chr9 | 16629206 | 16629207 | 1 | |
| Deletion | Chr3 | 17707732 | 17707733 | 1 | |
| Deletion | Chr4 | 18925440 | 18925444 | 4 | |
| Deletion | Chr1 | 21557001 | 21769000 | 211999 | 39 |
| Deletion | Chr1 | 21773001 | 21829000 | 55999 | 8 |
| Deletion | Chr11 | 24090093 | 24090094 | 1 | LOC_Os11g40410 |
| Deletion | Chr8 | 27502646 | 27502647 | 1 | |
| Deletion | Chr2 | 28276426 | 28276433 | 7 | |
| Deletion | Chr3 | 31604617 | 31604631 | 14 | |
| Deletion | Chr4 | 34957044 | 34957047 | 3 | |
| Deletion | Chr1 | 36302666 | 36302667 | 1 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr11 | 25831477 | 25831482 | 6 | |
| Insertion | Chr4 | 23097551 | 23097556 | 6 | |
| Insertion | Chr9 | 21903222 | 21903223 | 2 |
Inversions: 20
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr2 | 198255 | 198469 | |
| Inversion | Chr3 | 698722 | 698893 | LOC_Os03g02150 |
| Inversion | Chr12 | 1135559 | 1135742 | |
| Inversion | Chr10 | 1362810 | 1362943 | |
| Inversion | Chr5 | 1613989 | 1614166 | |
| Inversion | Chr2 | 2874264 | 2874530 | |
| Inversion | Chr5 | 2881057 | 2881173 | LOC_Os05g05780 |
| Inversion | Chr5 | 4115148 | 4115402 | |
| Inversion | Chr4 | 4646341 | 4646449 | |
| Inversion | Chr1 | 10067682 | 10067868 | |
| Inversion | Chr12 | 11400775 | 11400940 | |
| Inversion | Chr1 | 12874853 | 12874988 | |
| Inversion | Chr10 | 18270169 | 18646670 | LOC_Os10g34230 |
| Inversion | Chr10 | 18273764 | 18646675 | LOC_Os10g34230 |
| Inversion | Chr6 | 23669614 | 23669820 | |
| Inversion | Chr4 | 25728872 | 25728980 | LOC_Os04g43490 |
| Inversion | Chr7 | 26245040 | 26245144 | LOC_Os07g43890 |
| Inversion | Chr4 | 32223314 | 32223441 | LOC_Os04g54070 |
| Inversion | Chr1 | 39291070 | 39881959 | 2 |
| Inversion | Chr1 | 39291081 | 39881968 | 2 |
Translocations: 7
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 6312000 | Chr4 | 10315830 | |
| Translocation | Chr8 | 12316474 | Chr1 | 9101596 | 2 |
| Translocation | Chr8 | 13684513 | Chr4 | 10213111 | 2 |
| Translocation | Chr10 | 18270190 | Chr3 | 1090924 | 2 |
| Translocation | Chr10 | 18273744 | Chr3 | 1090843 | 2 |
| Translocation | Chr7 | 20170939 | Chr6 | 5157867 | |
| Translocation | Chr11 | 27840158 | Chr2 | 16105683 |
