Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2562-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2562-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 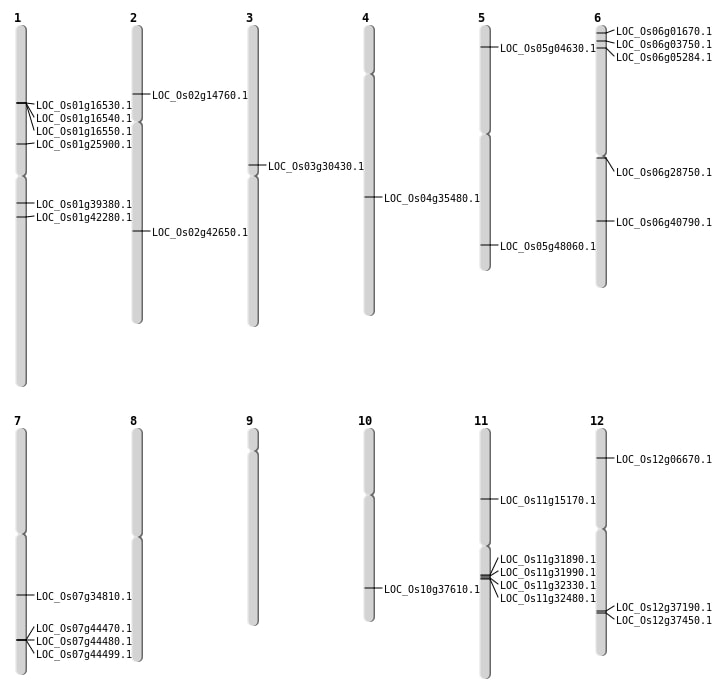
|
| Variant Information |
|---|
Single base substitutions: 42
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 39536349 | G-A | HET | ||
| SBS | Chr1 | 4733220 | C-T | HET | ||
| SBS | Chr1 | 9632093 | C-G | Homo | ||
| SBS | Chr10 | 9899038 | T-C | Homo | ||
| SBS | Chr11 | 10196191 | T-G | HET | ||
| SBS | Chr11 | 11724839 | G-A | HET | ||
| SBS | Chr11 | 19669103 | A-G | HET | ||
| SBS | Chr11 | 8551222 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g15170 |
| SBS | Chr12 | 16324591 | T-A | HET | ||
| SBS | Chr12 | 21002807 | C-G | HET | ||
| SBS | Chr12 | 22985686 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os12g37450 |
| SBS | Chr12 | 3243332 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os12g06670 |
| SBS | Chr2 | 33350716 | G-A | HET | ||
| SBS | Chr2 | 8165973 | A-C | HET | NON_SYNONYMOUS_CODING | LOC_Os02g14760 |
| SBS | Chr3 | 15533884 | C-T | Homo | ||
| SBS | Chr3 | 17348492 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os03g30430 |
| SBS | Chr3 | 22178010 | A-T | HET | ||
| SBS | Chr3 | 26897280 | C-T | HET | ||
| SBS | Chr4 | 14223449 | T-C | HET | ||
| SBS | Chr5 | 15947729 | C-T | HET | ||
| SBS | Chr5 | 24259200 | T-G | HET | ||
| SBS | Chr6 | 14013392 | T-A | HET | ||
| SBS | Chr6 | 1481258 | A-G | Homo | ||
| SBS | Chr6 | 15109188 | G-A | HET | ||
| SBS | Chr6 | 16374435 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os06g28750 |
| SBS | Chr6 | 17426312 | C-G | HET | ||
| SBS | Chr6 | 22200893 | T-C | HET | ||
| SBS | Chr6 | 22661402 | T-A | Homo | ||
| SBS | Chr6 | 8100871 | A-T | HET | ||
| SBS | Chr7 | 10267466 | T-A | HET | ||
| SBS | Chr7 | 20859206 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g34810 |
| SBS | Chr7 | 6535035 | A-G | HET | ||
| SBS | Chr8 | 16585240 | C-T | HET | ||
| SBS | Chr8 | 16585241 | C-T | HET | ||
| SBS | Chr8 | 16585242 | A-G | HET | ||
| SBS | Chr8 | 18632145 | G-T | HET | ||
| SBS | Chr8 | 23253605 | G-A | HET | ||
| SBS | Chr8 | 23953267 | T-C | HET | ||
| SBS | Chr8 | 25954157 | A-C | HET | ||
| SBS | Chr9 | 16875754 | G-A | HET | ||
| SBS | Chr9 | 17283553 | C-T | HET | ||
| SBS | Chr9 | 4620388 | T-A | HET |
Deletions: 21
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr1 | 452281 | 452301 | 20 | |
| Deletion | Chr10 | 1569997 | 1570005 | 8 | |
| Deletion | Chr3 | 4973989 | 4973990 | 1 | |
| Deletion | Chr8 | 8050583 | 8050589 | 6 | |
| Deletion | Chr5 | 9239325 | 9239333 | 8 | |
| Deletion | Chr1 | 9383001 | 9395000 | 11999 | 3 |
| Deletion | Chr11 | 12488241 | 12488307 | 66 | |
| Deletion | Chr9 | 13731582 | 13731587 | 5 | |
| Deletion | Chr3 | 17801528 | 17801541 | 13 | |
| Deletion | Chr3 | 18097368 | 18097382 | 14 | |
| Deletion | Chr4 | 18645066 | 18645074 | 8 | |
| Deletion | Chr10 | 20126496 | 20126502 | 6 | LOC_Os10g37610 |
| Deletion | Chr7 | 20949814 | 20949815 | 1 | |
| Deletion | Chr7 | 20983879 | 20983885 | 6 | |
| Deletion | Chr5 | 21162834 | 21162839 | 5 | |
| Deletion | Chr12 | 21847224 | 21847230 | 6 | |
| Deletion | Chr12 | 22807503 | 22807507 | 4 | LOC_Os12g37190 |
| Deletion | Chr12 | 23507074 | 23507079 | 5 | |
| Deletion | Chr7 | 26560001 | 26575000 | 14999 | 3 |
| Deletion | Chr6 | 30863486 | 30863487 | 1 | |
| Deletion | Chr2 | 31354246 | 31354263 | 17 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 19331298 | 19331299 | 2 | |
| Insertion | Chr4 | 21592832 | 21592832 | 1 | LOC_Os04g35480 |
| Insertion | Chr4 | 25528758 | 25528758 | 1 | |
| Insertion | Chr7 | 17560179 | 17560180 | 2 |
Inversions: 18
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr6 | 402710 | 402860 | LOC_Os06g01670 |
| Inversion | Chr6 | 1480521 | 2381886 | 2 |
| Inversion | Chr6 | 1480525 | 2381904 | 2 |
| Inversion | Chr5 | 2348565 | 2419793 | |
| Inversion | Chr1 | 4577769 | 4578016 | |
| Inversion | Chr5 | 6980346 | 6980570 | |
| Inversion | Chr4 | 10422512 | 10422617 | |
| Inversion | Chr7 | 15272352 | 15272467 | |
| Inversion | Chr11 | 18744799 | 18745017 | LOC_Os11g31890 |
| Inversion | Chr11 | 18849344 | 19096379 | 2 |
| Inversion | Chr11 | 19096390 | 19173182 | 2 |
| Inversion | Chr8 | 20641518 | 20641647 | |
| Inversion | Chr1 | 22163654 | 22163840 | LOC_Os01g39380 |
| Inversion | Chr1 | 23971288 | 23971553 | LOC_Os01g42280 |
| Inversion | Chr6 | 24337976 | 24338134 | LOC_Os06g40790 |
| Inversion | Chr2 | 25659240 | 25659345 | LOC_Os02g42650 |
| Inversion | Chr5 | 27557932 | 27558200 | LOC_Os05g48060 |
| Inversion | Chr4 | 35358223 | 35358332 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr5 | 2176739 | Chr1 | 14694252 | 2 |
| Translocation | Chr5 | 2285849 | Chr1 | 14694461 | 2 |
