Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2647-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2647-S Alignment File |
| Seed Availability | No |
| Mapping (Hover to Zoom-In) | 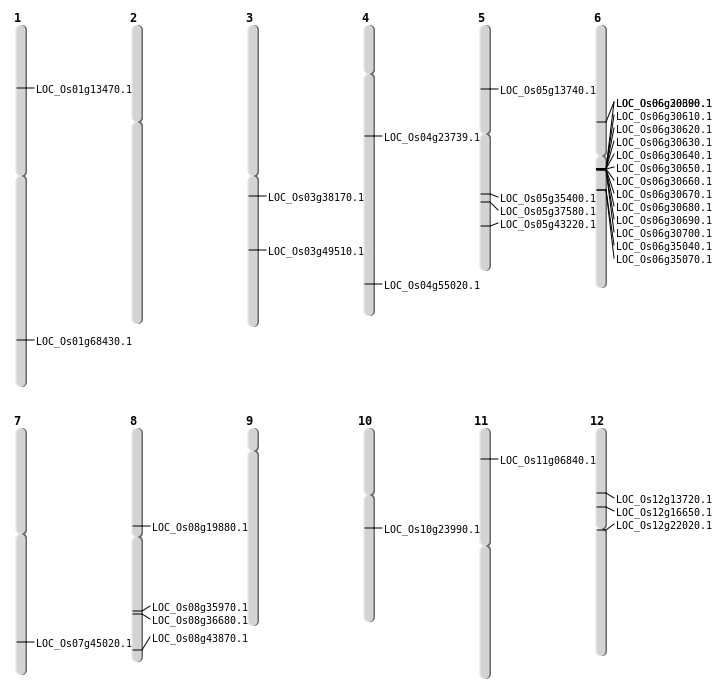
|
| Variant Information |
|---|
Single base substitutions: 106
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 14627508 | G-A | HET | INTRON | |
| SBS | Chr1 | 15768827 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 26325877 | T-C | HET | INTERGENIC | |
| SBS | Chr1 | 35732817 | G-A | HET | UTR_5_PRIME | |
| SBS | Chr1 | 3958249 | A-G | HET | UTR_5_PRIME | |
| SBS | Chr1 | 39768623 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os01g68430 |
| SBS | Chr1 | 7507478 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os01g13470 |
| SBS | Chr1 | 8342436 | T-C | HET | INTERGENIC | |
| SBS | Chr10 | 1181898 | G-T | HET | INTERGENIC | |
| SBS | Chr10 | 12115634 | C-A | HET | INTRON | |
| SBS | Chr10 | 12814949 | A-G | HOMO | INTERGENIC | |
| SBS | Chr10 | 18335018 | G-A | HET | INTRON | |
| SBS | Chr10 | 19950463 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 20981198 | A-G | HET | INTERGENIC | |
| SBS | Chr10 | 305090 | A-T | HET | INTRON | |
| SBS | Chr10 | 5756868 | T-A | HOMO | INTERGENIC | |
| SBS | Chr10 | 5893123 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 1005502 | A-G | HET | INTERGENIC | |
| SBS | Chr11 | 13954011 | G-T | HET | INTERGENIC | |
| SBS | Chr11 | 15389280 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 21477980 | T-C | HET | INTRON | |
| SBS | Chr11 | 21531714 | C-A | HET | INTERGENIC | |
| SBS | Chr11 | 3357703 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g06840 |
| SBS | Chr12 | 12386869 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os12g22020 |
| SBS | Chr12 | 14358316 | A-C | HOMO | INTERGENIC | |
| SBS | Chr12 | 15053756 | T-C | HET | INTERGENIC | |
| SBS | Chr12 | 21631046 | C-G | HOMO | INTERGENIC | |
| SBS | Chr12 | 24866285 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 24885607 | T-C | HET | INTERGENIC | |
| SBS | Chr12 | 7736618 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os12g13720 |
| SBS | Chr12 | 9534073 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os12g16650 |
| SBS | Chr2 | 19272436 | G-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 29797546 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 33123730 | G-T | HET | INTERGENIC | |
| SBS | Chr2 | 9970951 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 19682371 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 21198438 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os03g38170 |
| SBS | Chr3 | 22569038 | A-T | HET | INTERGENIC | |
| SBS | Chr3 | 23449384 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 26017064 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 28182656 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g49510 |
| SBS | Chr3 | 30329537 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 30951131 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 36376904 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr3 | 891335 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 9910661 | G-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 13572941 | A-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os04g23739 |
| SBS | Chr4 | 13572942 | A-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr4 | 1413254 | C-A | HET | INTERGENIC | |
| SBS | Chr4 | 16122930 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 16318392 | A-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 1746738 | G-T | HET | INTERGENIC | |
| SBS | Chr4 | 32713299 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g55020 |
| SBS | Chr4 | 33536468 | G-T | HET | INTERGENIC | |
| SBS | Chr4 | 33765070 | G-A | HET | UTR_3_PRIME | |
| SBS | Chr4 | 35331886 | T-C | HET | INTRON | |
| SBS | Chr4 | 5770799 | G-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 6606840 | C-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr4 | 7868226 | A-G | HET | INTERGENIC | |
| SBS | Chr5 | 13623931 | A-G | HET | INTRON | |
| SBS | Chr5 | 17005326 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 18324433 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 21034196 | T-C | HOMO | NON_SYNONYMOUS_CODING | LOC_Os05g35400 |
| SBS | Chr5 | 2183723 | C-T | HET | INTRON | |
| SBS | Chr5 | 21990369 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g37580 |
| SBS | Chr5 | 25474599 | T-A | HET | INTERGENIC | |
| SBS | Chr5 | 7616070 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g13740 |
| SBS | Chr5 | 9954684 | G-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 10742687 | T-C | HET | INTERGENIC | |
| SBS | Chr6 | 11716334 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os06g20390 |
| SBS | Chr6 | 12869957 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr6 | 13269268 | T-C | HET | INTERGENIC | |
| SBS | Chr6 | 17532837 | C-A | HET | INTERGENIC | |
| SBS | Chr6 | 20394192 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os06g35040 |
| SBS | Chr6 | 21653431 | G-T | HET | INTERGENIC | |
| SBS | Chr6 | 23881583 | A-T | HET | INTERGENIC | |
| SBS | Chr6 | 6547923 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 7176349 | C-A | HET | INTERGENIC | |
| SBS | Chr7 | 10913939 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr7 | 11449826 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 17435436 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 18357799 | C-G | HET | INTERGENIC | |
| SBS | Chr7 | 20357082 | A-T | HET | INTERGENIC | |
| SBS | Chr7 | 26867327 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g45020 |
| SBS | Chr7 | 646692 | G-T | HOMO | INTRON | |
| SBS | Chr7 | 837814 | T-A | HET | INTERGENIC | |
| SBS | Chr8 | 11910550 | C-A | HET | STOP_GAINED | LOC_Os08g19880 |
| SBS | Chr8 | 12852078 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 15584835 | C-G | HET | INTERGENIC | |
| SBS | Chr8 | 17059792 | C-G | HET | INTERGENIC | |
| SBS | Chr8 | 18080379 | G-T | HET | INTERGENIC | |
| SBS | Chr8 | 19048950 | A-G | HET | SYNONYMOUS_CODING | |
| SBS | Chr8 | 21865655 | C-G | HET | INTERGENIC | |
| SBS | Chr8 | 2202130 | A-T | HET | INTERGENIC | |
| SBS | Chr8 | 23148918 | T-A | HET | STOP_GAINED | LOC_Os08g36680 |
| SBS | Chr8 | 23825145 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 5470003 | C-A | HET | INTERGENIC | |
| SBS | Chr9 | 11624397 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr9 | 13941064 | A-G | HET | INTERGENIC | |
| SBS | Chr9 | 15668747 | T-C | HET | INTERGENIC | |
| SBS | Chr9 | 15668751 | T-A | HET | INTERGENIC | |
| SBS | Chr9 | 15668755 | G-T | HET | INTERGENIC | |
| SBS | Chr9 | 21976012 | A-T | HOMO | INTERGENIC | |
| SBS | Chr9 | 22067407 | G-A | HOMO | INTRON | |
| SBS | Chr9 | 22211832 | C-G | HOMO | INTERGENIC | |
| SBS | Chr9 | 8007969 | G-A | HET | SYNONYMOUS_CODING |
Deletions: 29
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr10 | 921349 | 921350 | 1 | |
| Deletion | Chr11 | 4697643 | 4697644 | 1 | |
| Deletion | Chr10 | 6227446 | 6227462 | 16 | |
| Deletion | Chr9 | 6621135 | 6621151 | 16 | |
| Deletion | Chr9 | 7848526 | 7848533 | 7 | |
| Deletion | Chr9 | 8377464 | 8377468 | 4 | |
| Deletion | Chr8 | 9964208 | 9964210 | 2 | |
| Deletion | Chr6 | 10272186 | 10272194 | 8 | |
| Deletion | Chr11 | 10543036 | 10543038 | 2 | |
| Deletion | Chr6 | 10723124 | 10723125 | 1 | |
| Deletion | Chr10 | 12291178 | 12291179 | 1 | LOC_Os10g23990 |
| Deletion | Chr11 | 13249491 | 13249494 | 3 | |
| Deletion | Chr6 | 13798969 | 13798970 | 1 | |
| Deletion | Chr2 | 15223241 | 15223254 | 13 | |
| Deletion | Chr10 | 17070057 | 17070065 | 8 | |
| Deletion | Chr4 | 17370205 | 17370209 | 4 | |
| Deletion | Chr6 | 17698001 | 17799000 | 100999 | 12 |
| Deletion | Chr8 | 19009281 | 19009293 | 12 | |
| Deletion | Chr4 | 19080774 | 19080784 | 10 | |
| Deletion | Chr6 | 19917114 | 19917120 | 6 | |
| Deletion | Chr5 | 20603982 | 20603998 | 16 | |
| Deletion | Chr3 | 22273445 | 22273447 | 2 | |
| Deletion | Chr10 | 22953981 | 22953982 | 1 | |
| Deletion | Chr5 | 25134414 | 25134416 | 2 | LOC_Os05g43220 |
| Deletion | Chr6 | 26367669 | 26367677 | 8 | |
| Deletion | Chr8 | 27670187 | 27670195 | 8 | LOC_Os08g43870 |
| Deletion | Chr5 | 28022850 | 28022855 | 5 | |
| Deletion | Chr4 | 29077100 | 29077105 | 5 | |
| Deletion | Chr2 | 30158869 | 30158895 | 26 |
Insertions: 13
Inversions: 6
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr10 | 14363390 | Chr8 | 27766504 | |
| Translocation | Chr10 | 14363404 | Chr8 | 27768594 | |
| Translocation | Chr9 | 16540012 | Chr6 | 17497729 | |
| Translocation | Chr8 | 22680582 | Chr7 | 23002853 | LOC_Os08g35970 |
| Translocation | Chr8 | 27265369 | Chr1 | 18068368 |
