Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2648-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2648-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 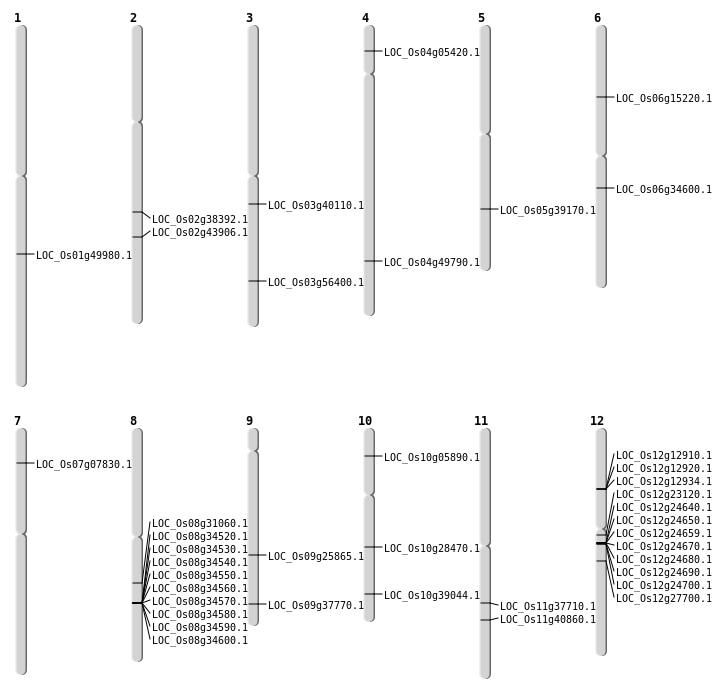
|
| Variant Information |
|---|
Single base substitutions: 54
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 14665619 | G-A | HET | ||
| SBS | Chr1 | 22729856 | C-T | Homo | ||
| SBS | Chr1 | 30638613 | C-T | HET | ||
| SBS | Chr1 | 34260188 | T-C | HET | ||
| SBS | Chr1 | 8472791 | C-G | Homo | ||
| SBS | Chr10 | 14813865 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g28470 |
| SBS | Chr10 | 2972382 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os10g05890 |
| SBS | Chr10 | 7020206 | G-A | Homo | ||
| SBS | Chr10 | 7579442 | C-T | HET | ||
| SBS | Chr11 | 13825554 | G-C | HET | ||
| SBS | Chr11 | 25143238 | G-C | HET | ||
| SBS | Chr11 | 4318184 | A-C | HET | ||
| SBS | Chr12 | 10776927 | G-A | HET | ||
| SBS | Chr12 | 13074191 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g23120 |
| SBS | Chr12 | 14456304 | T-A | HET | ||
| SBS | Chr12 | 1774708 | C-T | HET | ||
| SBS | Chr12 | 9427676 | C-T | HET | ||
| SBS | Chr2 | 14689894 | C-G | HET | ||
| SBS | Chr2 | 15275271 | G-A | HET | ||
| SBS | Chr2 | 23214240 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os02g38392 |
| SBS | Chr2 | 25774240 | G-A | HET | ||
| SBS | Chr2 | 26507445 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os02g43906 |
| SBS | Chr2 | 26507452 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os02g43906 |
| SBS | Chr2 | 35714688 | G-C | Homo | ||
| SBS | Chr3 | 12233342 | A-G | Homo | ||
| SBS | Chr3 | 22292358 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os03g40110 |
| SBS | Chr3 | 32151790 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os03g56400 |
| SBS | Chr3 | 35663492 | G-A | HET | ||
| SBS | Chr4 | 12800103 | C-A | HET | ||
| SBS | Chr4 | 2427569 | C-T | HET | ||
| SBS | Chr4 | 24762958 | G-A | HET | ||
| SBS | Chr4 | 26150540 | G-A | Homo | ||
| SBS | Chr4 | 30427457 | G-A | HET | ||
| SBS | Chr5 | 10335794 | C-T | Homo | ||
| SBS | Chr5 | 12129925 | C-T | HET | ||
| SBS | Chr5 | 22971043 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g39170 |
| SBS | Chr5 | 27200736 | T-C | Homo | ||
| SBS | Chr5 | 28402682 | C-T | HET | ||
| SBS | Chr5 | 28808740 | C-T | HET | ||
| SBS | Chr5 | 29686691 | A-T | HET | ||
| SBS | Chr5 | 5594979 | C-T | HET | ||
| SBS | Chr6 | 16421327 | C-A | HET | ||
| SBS | Chr6 | 20121665 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os06g34600 |
| SBS | Chr6 | 5909972 | T-C | HET | ||
| SBS | Chr6 | 6002346 | T-C | HET | ||
| SBS | Chr7 | 2907825 | A-G | Homo | ||
| SBS | Chr7 | 4758388 | C-T | Homo | ||
| SBS | Chr7 | 9541200 | G-T | HET | ||
| SBS | Chr8 | 17094992 | G-A | Homo | ||
| SBS | Chr8 | 19175431 | G-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os08g31060 |
| SBS | Chr8 | 24110107 | T-C | HET | ||
| SBS | Chr8 | 28324506 | T-A | HET | ||
| SBS | Chr8 | 7184452 | C-A | HET | ||
| SBS | Chr8 | 7676779 | G-A | Homo |
Deletions: 32
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr7 | 1082946 | 1082975 | 29 | |
| Deletion | Chr4 | 2061693 | 2061695 | 2 | |
| Deletion | Chr12 | 2504327 | 2504332 | 5 | |
| Deletion | Chr9 | 4753131 | 4753204 | 73 | |
| Deletion | Chr12 | 7148001 | 7168000 | 19999 | 3 |
| Deletion | Chr1 | 7813719 | 7813721 | 2 | |
| Deletion | Chr6 | 8622641 | 8622642 | 1 | LOC_Os06g15220 |
| Deletion | Chr2 | 9139505 | 9139653 | 148 | |
| Deletion | Chr2 | 9448349 | 9448350 | 1 | |
| Deletion | Chr5 | 10873678 | 10873689 | 11 | |
| Deletion | Chr1 | 12473007 | 12473008 | 1 | |
| Deletion | Chr3 | 12666713 | 12666717 | 4 | |
| Deletion | Chr12 | 14102001 | 14165000 | 62999 | 7 |
| Deletion | Chr12 | 14137706 | 14137707 | 1 | |
| Deletion | Chr4 | 16244040 | 16244041 | 1 | |
| Deletion | Chr10 | 17414700 | 17414702 | 2 | |
| Deletion | Chr6 | 17684927 | 17685052 | 125 | |
| Deletion | Chr5 | 17855227 | 17855229 | 2 | |
| Deletion | Chr7 | 20887274 | 20887276 | 2 | |
| Deletion | Chr8 | 21664001 | 21747000 | 82999 | 9 |
| Deletion | Chr9 | 21759868 | 21759874 | 6 | LOC_Os09g37770 |
| Deletion | Chr11 | 22293402 | 22293403 | 1 | LOC_Os11g37710 |
| Deletion | Chr11 | 23956730 | 23956731 | 1 | |
| Deletion | Chr5 | 25108743 | 25108745 | 2 | |
| Deletion | Chr12 | 25183530 | 25183553 | 23 | |
| Deletion | Chr4 | 25268617 | 25268625 | 8 | |
| Deletion | Chr1 | 26245482 | 26245484 | 2 | |
| Deletion | Chr4 | 27284930 | 27284931 | 1 | |
| Deletion | Chr1 | 28721690 | 28721732 | 42 | LOC_Os01g49980 |
| Deletion | Chr1 | 31339955 | 31339962 | 7 | |
| Deletion | Chr1 | 31355889 | 31355890 | 1 | |
| Deletion | Chr2 | 35385679 | 35385681 | 2 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 33577823 | 33577824 | 2 | |
| Insertion | Chr12 | 3582702 | 3582703 | 2 | |
| Insertion | Chr7 | 106866 | 106887 | 22 |
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr4 | 2715364 | 2783946 | LOC_Os04g05420 |
| Inversion | Chr4 | 2715417 | 2783955 | LOC_Os04g05420 |
| Inversion | Chr9 | 14587791 | 15506068 | 2 |
| Inversion | Chr9 | 14587798 | 15506075 | 2 |
| Inversion | Chr2 | 27431306 | 27501701 |
Translocations: 4
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 16335307 | Chr7 | 3932772 | 2 |
| Translocation | Chr4 | 17221223 | Chr3 | 25790868 | |
| Translocation | Chr11 | 24445386 | Chr4 | 29686988 | 2 |
| Translocation | Chr11 | 26331021 | Chr10 | 20813219 | 2 |
