Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2649-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2649-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 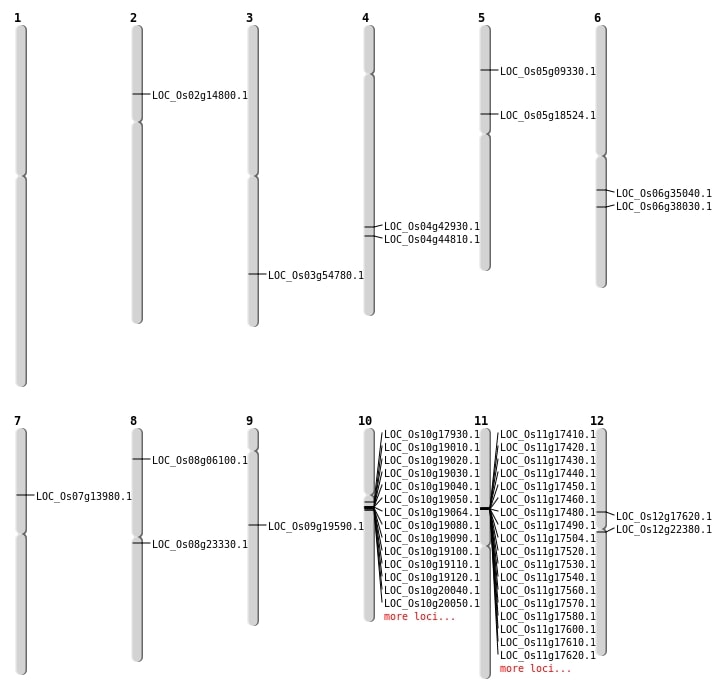
|
| Variant Information |
|---|
Single base substitutions: 70
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 13336061 | G-A | HET | ||
| SBS | Chr1 | 176284 | C-T | HET | ||
| SBS | Chr1 | 25500539 | C-A | HET | ||
| SBS | Chr1 | 31822101 | C-A | HET | ||
| SBS | Chr1 | 32310095 | C-T | HET | ||
| SBS | Chr1 | 37317060 | C-T | HET | ||
| SBS | Chr1 | 40882386 | T-A | HET | ||
| SBS | Chr1 | 40882387 | C-A | HET | ||
| SBS | Chr1 | 6677786 | T-A | HET | ||
| SBS | Chr10 | 1338387 | G-T | HET | ||
| SBS | Chr10 | 17850953 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os10g33760 |
| SBS | Chr10 | 23174384 | A-G | HET | ||
| SBS | Chr10 | 8159402 | T-C | HET | ||
| SBS | Chr11 | 1128861 | C-T | HET | ||
| SBS | Chr11 | 1749366 | T-C | HET | ||
| SBS | Chr11 | 20154989 | T-C | Homo | ||
| SBS | Chr11 | 20313274 | G-A | HET | ||
| SBS | Chr11 | 3108154 | C-A | HET | ||
| SBS | Chr11 | 3345895 | T-G | HET | ||
| SBS | Chr12 | 10092256 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g17620 |
| SBS | Chr12 | 12640022 | T-G | Homo | NON_SYNONYMOUS_CODING | LOC_Os12g22380 |
| SBS | Chr12 | 15659424 | A-G | HET | ||
| SBS | Chr12 | 18744990 | C-T | HET | ||
| SBS | Chr12 | 8977088 | G-A | HET | ||
| SBS | Chr2 | 24817503 | A-G | Homo | ||
| SBS | Chr2 | 25577667 | A-T | Homo | ||
| SBS | Chr2 | 27527420 | T-G | HET | ||
| SBS | Chr2 | 27527421 | G-A | HET | ||
| SBS | Chr2 | 27527423 | G-A | HET | ||
| SBS | Chr2 | 27541766 | C-G | HET | ||
| SBS | Chr2 | 28773698 | T-A | HET | ||
| SBS | Chr2 | 30060404 | C-T | HET | ||
| SBS | Chr3 | 10402584 | C-T | Homo | ||
| SBS | Chr3 | 15388459 | G-A | HET | ||
| SBS | Chr3 | 22362037 | C-T | HET | ||
| SBS | Chr3 | 27760967 | C-A | HET | ||
| SBS | Chr3 | 28370256 | C-T | HET | ||
| SBS | Chr3 | 31140188 | C-T | HET | ||
| SBS | Chr3 | 31140189 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g54780 |
| SBS | Chr3 | 6141836 | G-A | HET | ||
| SBS | Chr3 | 9532892 | G-A | HET | ||
| SBS | Chr4 | 17075214 | T-A | HET | ||
| SBS | Chr4 | 17599973 | T-A | Homo | ||
| SBS | Chr4 | 25207369 | C-T | HET | ||
| SBS | Chr4 | 557185 | A-G | HET | ||
| SBS | Chr5 | 1679366 | C-A | HET | ||
| SBS | Chr5 | 20469569 | T-A | HET | ||
| SBS | Chr5 | 22717538 | G-T | HET | ||
| SBS | Chr5 | 29621611 | C-G | Homo | ||
| SBS | Chr5 | 8964460 | G-A | HET | ||
| SBS | Chr5 | 9128311 | T-C | HET | ||
| SBS | Chr6 | 20394439 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os06g35040 |
| SBS | Chr6 | 2690082 | A-G | HET | ||
| SBS | Chr6 | 29874744 | C-T | HET | ||
| SBS | Chr7 | 14967868 | C-T | HET | ||
| SBS | Chr7 | 16393472 | G-A | HET | ||
| SBS | Chr7 | 28933587 | G-A | HET | ||
| SBS | Chr7 | 6061405 | A-T | HET | ||
| SBS | Chr7 | 9440791 | A-G | HET | ||
| SBS | Chr8 | 13007215 | T-C | HET | ||
| SBS | Chr8 | 13591919 | C-A | HET | ||
| SBS | Chr8 | 2500913 | C-T | HET | ||
| SBS | Chr8 | 3338215 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g06100 |
| SBS | Chr8 | 3338216 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g06100 |
| SBS | Chr9 | 11717057 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os09g19590 |
| SBS | Chr9 | 14964326 | G-A | HET | ||
| SBS | Chr9 | 17300471 | G-C | HET | ||
| SBS | Chr9 | 17535915 | C-A | HET | ||
| SBS | Chr9 | 22050975 | G-C | HET | ||
| SBS | Chr9 | 2425367 | C-T | Homo |
Deletions: 15
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr11 | 1198931 | 1198932 | 1 | |
| Deletion | Chr11 | 2290254 | 2290255 | 1 | |
| Deletion | Chr10 | 9063321 | 9063322 | 1 | LOC_Os10g17930 |
| Deletion | Chr10 | 9682001 | 9734000 | 51999 | 11 |
| Deletion | Chr11 | 9707001 | 9999000 | 291999 | 38 |
| Deletion | Chr10 | 10056001 | 10189000 | 132999 | 20 |
| Deletion | Chr10 | 11052488 | 11052489 | 1 | |
| Deletion | Chr2 | 11384599 | 11384603 | 4 | |
| Deletion | Chr8 | 20140398 | 20140406 | 8 | |
| Deletion | Chr6 | 22481456 | 22481460 | 4 | LOC_Os06g38030 |
| Deletion | Chr4 | 26134270 | 26134287 | 17 | |
| Deletion | Chr6 | 26606594 | 26606603 | 9 | |
| Deletion | Chr5 | 27338343 | 27338348 | 5 | |
| Deletion | Chr1 | 28566624 | 28566625 | 1 | |
| Deletion | Chr2 | 33476510 | 33476512 | 2 |
Insertions: 5
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr2 | 7970279 | 8211576 | 2 |
| Inversion | Chr2 | 7970293 | 8238782 |
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr5 | 10722802 | Chr4 | 26530140 | 2 |
| Translocation | Chr11 | 11737981 | Chr4 | 25397688 | 2 |
| Translocation | Chr11 | 11749922 | Chr4 | 25397690 | 2 |
| Translocation | Chr8 | 14086287 | Chr5 | 5212205 | 2 |
| Translocation | Chr11 | 21589576 | Chr7 | 7985005 | 2 |
