Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2654-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2654-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 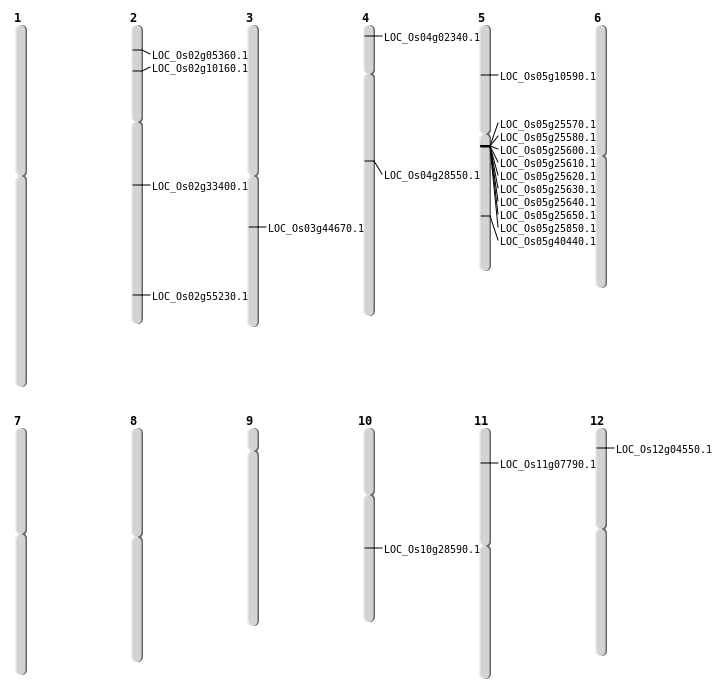
|
| Variant Information |
|---|
Single base substitutions: 50
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 11859923 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 13337197 | T-A | HET | INTERGENIC | |
| SBS | Chr1 | 14230251 | A-T | HET | INTRON | |
| SBS | Chr1 | 17041475 | T-A | HET | INTRON | |
| SBS | Chr1 | 22060509 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 3975927 | G-A | HET | UTR_5_PRIME | |
| SBS | Chr1 | 5671382 | C-T | HET | UTR_5_PRIME | |
| SBS | Chr10 | 16096311 | T-C | HET | INTRON | |
| SBS | Chr10 | 16647213 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 17357182 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 3176816 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 3869787 | T-G | HET | UTR_3_PRIME | |
| SBS | Chr10 | 8683184 | C-T | HET | INTRON | |
| SBS | Chr10 | 9698715 | T-C | HET | INTERGENIC | |
| SBS | Chr10 | 9698716 | A-G | HET | INTERGENIC | |
| SBS | Chr11 | 1617203 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr11 | 19655329 | C-G | HET | INTRON | |
| SBS | Chr11 | 20050670 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 10047192 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 16757674 | G-A | HET | INTRON | |
| SBS | Chr12 | 1933607 | A-C | HET | NON_SYNONYMOUS_CODING | LOC_Os12g04550 |
| SBS | Chr12 | 23853668 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 26802803 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 13273669 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 2576372 | C-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os02g05360 |
| SBS | Chr2 | 5425094 | A-G | HOMO | INTERGENIC | |
| SBS | Chr2 | 6770951 | A-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 27245734 | C-T | HET | INTRON | |
| SBS | Chr4 | 11152782 | G-T | HET | INTERGENIC | |
| SBS | Chr4 | 16021497 | C-A | HET | INTERGENIC | |
| SBS | Chr4 | 16906017 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g28550 |
| SBS | Chr4 | 22323938 | C-G | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr4 | 6211041 | C-A | HET | INTERGENIC | |
| SBS | Chr4 | 8251075 | C-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 18951325 | A-G | HET | INTERGENIC | |
| SBS | Chr5 | 2439330 | A-C | HET | INTERGENIC | |
| SBS | Chr5 | 24914904 | A-G | HET | INTERGENIC | |
| SBS | Chr5 | 27801535 | T-C | HET | INTRON | |
| SBS | Chr5 | 5775765 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 11953211 | A-G | HET | INTRON | |
| SBS | Chr6 | 28383275 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 6297141 | T-C | HET | INTRON | |
| SBS | Chr6 | 6297149 | G-A | HET | INTRON | |
| SBS | Chr8 | 11868671 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 12188597 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 14471600 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 21876495 | A-T | HET | INTRON | |
| SBS | Chr9 | 21876498 | A-G | HET | INTRON | |
| SBS | Chr9 | 9658128 | C-G | HOMO | INTRON | |
| SBS | Chr9 | 9658135 | A-G | HOMO | INTRON |
Deletions: 34
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr2 | 213994 | 213999 | 5 | |
| Deletion | Chr4 | 821152 | 821225 | 73 | LOC_Os04g02340 |
| Deletion | Chr7 | 1480786 | 1480787 | 1 | |
| Deletion | Chr6 | 1710560 | 1710561 | 1 | |
| Deletion | Chr3 | 2766382 | 2766416 | 34 | |
| Deletion | Chr1 | 3059980 | 3059982 | 2 | |
| Deletion | Chr11 | 3971118 | 3971122 | 4 | LOC_Os11g07790 |
| Deletion | Chr6 | 4745706 | 4745709 | 3 | |
| Deletion | Chr2 | 5326961 | 5326972 | 11 | LOC_Os02g10160 |
| Deletion | Chr3 | 5617505 | 5617515 | 10 | |
| Deletion | Chr6 | 5751858 | 5751860 | 2 | |
| Deletion | Chr6 | 5752189 | 5752195 | 6 | |
| Deletion | Chr3 | 9339504 | 9339528 | 24 | |
| Deletion | Chr8 | 10774837 | 10774838 | 1 | |
| Deletion | Chr9 | 11396116 | 11396127 | 11 | |
| Deletion | Chr10 | 11705248 | 11705252 | 4 | |
| Deletion | Chr7 | 12872595 | 12872596 | 1 | |
| Deletion | Chr5 | 14853001 | 14909000 | 55999 | 9 |
| Deletion | Chr10 | 14889519 | 14889522 | 3 | LOC_Os10g28590 |
| Deletion | Chr7 | 17002361 | 17002369 | 8 | |
| Deletion | Chr1 | 18277124 | 18277196 | 72 | |
| Deletion | Chr2 | 19844275 | 19844277 | 2 | LOC_Os02g33400 |
| Deletion | Chr10 | 19937088 | 19937090 | 2 | |
| Deletion | Chr5 | 22987841 | 22987844 | 3 | |
| Deletion | Chr5 | 23758506 | 23758511 | 5 | LOC_Os05g40440 |
| Deletion | Chr7 | 24474030 | 24474035 | 5 | |
| Deletion | Chr6 | 24873024 | 24873025 | 1 | |
| Deletion | Chr1 | 26253062 | 26253068 | 6 | |
| Deletion | Chr3 | 27783836 | 27783842 | 6 | |
| Deletion | Chr8 | 28150708 | 28150710 | 2 | |
| Deletion | Chr5 | 29081508 | 29081514 | 6 | |
| Deletion | Chr3 | 33314658 | 33314665 | 7 | |
| Deletion | Chr2 | 33827389 | 33827392 | 3 | LOC_Os02g55230 |
| Deletion | Chr4 | 35285173 | 35285174 | 1 |
Insertions: 10
No Inversion
Translocations: 6
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr5 | 32916 | Chr3 | 23639478 | |
| Translocation | Chr9 | 7057099 | Chr1 | 31953042 | |
| Translocation | Chr11 | 17836748 | Chr10 | 12209849 | |
| Translocation | Chr11 | 22532682 | Chr5 | 5767275 | LOC_Os05g10590 |
| Translocation | Chr3 | 24883120 | Chr2 | 8852240 | |
| Translocation | Chr6 | 25585467 | Chr3 | 25151135 | LOC_Os03g44670 |
