Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2656-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2656-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 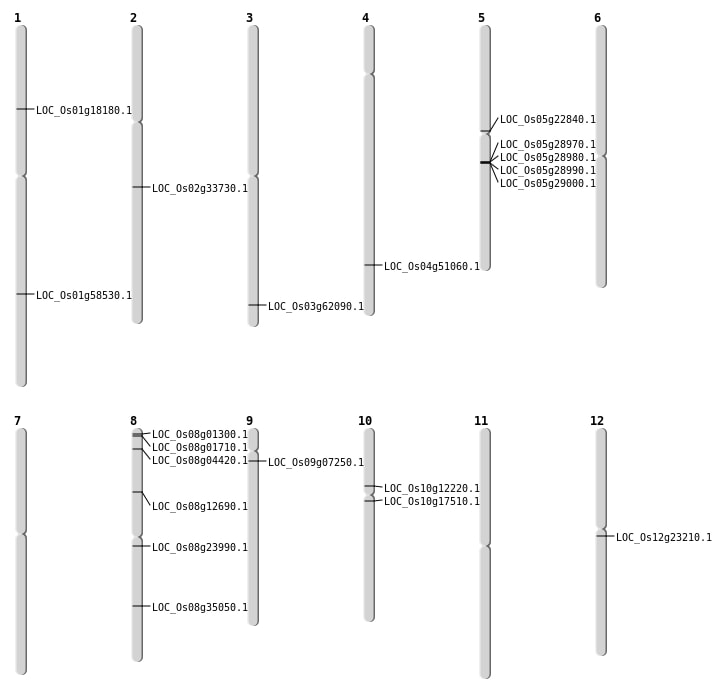
|
| Variant Information |
|---|
Single base substitutions: 49
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 4655561 | G-A | HET | ||
| SBS | Chr10 | 16546967 | C-T | Homo | ||
| SBS | Chr10 | 6803825 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g12220 |
| SBS | Chr11 | 15351998 | C-T | Homo | ||
| SBS | Chr11 | 16749525 | G-A | Homo | ||
| SBS | Chr11 | 19628155 | C-A | HET | ||
| SBS | Chr11 | 25284009 | C-T | HET | ||
| SBS | Chr11 | 26037426 | C-T | HET | ||
| SBS | Chr11 | 27195885 | T-A | Homo | ||
| SBS | Chr11 | 28176843 | A-T | HET | ||
| SBS | Chr11 | 9873874 | C-T | HET | ||
| SBS | Chr12 | 13120896 | C-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os12g23210 |
| SBS | Chr12 | 19206691 | G-A | HET | ||
| SBS | Chr12 | 26436879 | G-A | HET | ||
| SBS | Chr12 | 27161448 | A-G | HET | ||
| SBS | Chr2 | 21843589 | T-G | HET | ||
| SBS | Chr2 | 28389313 | T-C | HET | ||
| SBS | Chr2 | 9553036 | G-A | HET | ||
| SBS | Chr3 | 16939040 | G-A | Homo | ||
| SBS | Chr3 | 16939041 | A-T | Homo | ||
| SBS | Chr3 | 30099420 | G-C | Homo | ||
| SBS | Chr3 | 34514925 | A-G | HET | ||
| SBS | Chr3 | 35178962 | G-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os03g62090 |
| SBS | Chr3 | 35625244 | A-G | HET | ||
| SBS | Chr3 | 6166646 | A-T | Homo | ||
| SBS | Chr4 | 34215226 | G-T | Homo | ||
| SBS | Chr5 | 12959923 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g22840 |
| SBS | Chr5 | 17685142 | C-G | Homo | ||
| SBS | Chr5 | 25419405 | G-A | HET | ||
| SBS | Chr6 | 10487633 | G-A | HET | ||
| SBS | Chr6 | 11184329 | C-T | HET | ||
| SBS | Chr6 | 17500502 | A-T | HET | ||
| SBS | Chr6 | 23190926 | C-A | HET | ||
| SBS | Chr6 | 26487539 | C-T | HET | ||
| SBS | Chr6 | 26487542 | A-T | HET | ||
| SBS | Chr6 | 26707474 | A-T | HET | ||
| SBS | Chr7 | 10118511 | G-A | HET | ||
| SBS | Chr7 | 12618896 | C-T | HET | ||
| SBS | Chr7 | 15390130 | C-T | HET | ||
| SBS | Chr7 | 15414217 | T-C | HET | ||
| SBS | Chr7 | 28158777 | A-G | HET | ||
| SBS | Chr8 | 2160701 | C-T | HET | SPLICE_SITE_DONOR | LOC_Os08g04420 |
| SBS | Chr8 | 7521889 | T-C | Homo | NON_SYNONYMOUS_CODING | LOC_Os08g12690 |
| SBS | Chr9 | 1328513 | C-T | HET | ||
| SBS | Chr9 | 15644506 | T-A | HET | ||
| SBS | Chr9 | 17446565 | G-C | HET | ||
| SBS | Chr9 | 20613353 | A-C | HET | ||
| SBS | Chr9 | 2295382 | A-T | HET | ||
| SBS | Chr9 | 2295385 | T-C | HET |
Deletions: 27
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr11 | 644937 | 644950 | 13 | |
| Deletion | Chr1 | 1162213 | 1162224 | 11 | |
| Deletion | Chr11 | 1319881 | 1319891 | 10 | |
| Deletion | Chr9 | 1541936 | 1541951 | 15 | |
| Deletion | Chr12 | 2387976 | 2387993 | 17 | |
| Deletion | Chr11 | 2546014 | 2546018 | 4 | |
| Deletion | Chr9 | 2957495 | 2957498 | 3 | |
| Deletion | Chr12 | 3912336 | 3912339 | 3 | |
| Deletion | Chr7 | 4145216 | 4145230 | 14 | |
| Deletion | Chr10 | 8832527 | 8832528 | 1 | LOC_Os10g17510 |
| Deletion | Chr1 | 10177333 | 10177337 | 4 | LOC_Os01g18180 |
| Deletion | Chr5 | 11356865 | 11356878 | 13 | |
| Deletion | Chr6 | 12412543 | 12412544 | 1 | |
| Deletion | Chr6 | 13200104 | 13200105 | 1 | |
| Deletion | Chr5 | 16987001 | 16999000 | 11999 | 4 |
| Deletion | Chr10 | 19459804 | 19459806 | 2 | |
| Deletion | Chr2 | 20127525 | 20127531 | 6 | LOC_Os02g33730 |
| Deletion | Chr5 | 20410264 | 20410276 | 12 | |
| Deletion | Chr3 | 20754338 | 20754339 | 1 | |
| Deletion | Chr5 | 23314067 | 23314073 | 6 | |
| Deletion | Chr11 | 24201711 | 24201728 | 17 | |
| Deletion | Chr7 | 28838992 | 28839002 | 10 | |
| Deletion | Chr1 | 29646797 | 29646798 | 1 | |
| Deletion | Chr4 | 30227532 | 30227534 | 2 | LOC_Os04g51060 |
| Deletion | Chr3 | 31521370 | 31521374 | 4 | |
| Deletion | Chr4 | 32062697 | 32062698 | 1 | |
| Deletion | Chr3 | 35215054 | 35215056 | 2 |
Insertions: 2
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr2 | 4396816 | 4396817 | 2 | |
| Insertion | Chr3 | 23133210 | 23133211 | 2 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr9 | 3527050 | 3557421 | 2 |
| Inversion | Chr9 | 3527224 | 3558152 | 2 |
Translocations: 13
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr8 | 192323 | Chr7 | 4497657 | LOC_Os08g01300 |
| Translocation | Chr3 | 268159 | Chr2 | 29684992 | |
| Translocation | Chr3 | 268164 | Chr2 | 29684996 | |
| Translocation | Chr8 | 435452 | Chr7 | 4497647 | LOC_Os08g01710 |
| Translocation | Chr12 | 1816568 | Chr5 | 24756772 | |
| Translocation | Chr12 | 1816574 | Chr5 | 24756620 | |
| Translocation | Chr9 | 3557421 | Chr8 | 22088590 | 2 |
| Translocation | Chr9 | 3558145 | Chr8 | 22088605 | 2 |
| Translocation | Chr9 | 11307386 | Chr6 | 22141205 | |
| Translocation | Chr8 | 14513929 | Chr1 | 29953971 | LOC_Os08g23990 |
| Translocation | Chr8 | 14513930 | Chr1 | 29953040 | LOC_Os08g23990 |
| Translocation | Chr2 | 22470361 | Chr1 | 33825445 | 2 |
| Translocation | Chr2 | 22470367 | Chr1 | 33824808 | 2 |
