Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2657-S-1 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2657-S-1 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 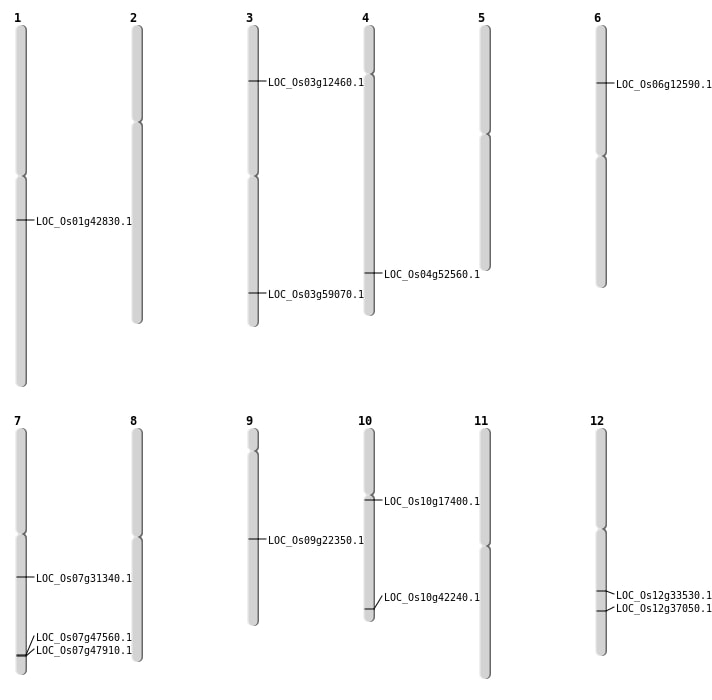
|
| Variant Information |
|---|
Single base substitutions: 67
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 12075898 | A-T | HET | INTERGENIC | |
| SBS | Chr1 | 12129599 | A-G | HET | UTR_3_PRIME | |
| SBS | Chr1 | 20150662 | T-C | HET | INTERGENIC | |
| SBS | Chr1 | 24115542 | G-C | HET | INTERGENIC | |
| SBS | Chr1 | 24377951 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g42830 |
| SBS | Chr1 | 3131828 | G-T | HET | INTERGENIC | |
| SBS | Chr1 | 40423792 | C-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 40793911 | A-G | HET | INTERGENIC | |
| SBS | Chr1 | 7031170 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 9560623 | C-G | HET | INTERGENIC | |
| SBS | Chr1 | 9856877 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 122153 | G-A | HET | INTRON | |
| SBS | Chr10 | 22751246 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g42240 |
| SBS | Chr10 | 4453900 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 17841574 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 14045077 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 16117761 | C-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 16184264 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr12 | 20260198 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os12g33530 |
| SBS | Chr12 | 22705168 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os12g37050 |
| SBS | Chr12 | 4407270 | C-A | HET | INTERGENIC | |
| SBS | Chr12 | 7817618 | G-C | HET | INTERGENIC | |
| SBS | Chr2 | 11652177 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 19517941 | C-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 35931594 | C-G | HOMO | #VALUE! | |
| SBS | Chr2 | 4752019 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 16652106 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 20569314 | C-A | HET | UTR_5_PRIME | |
| SBS | Chr3 | 24784694 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 29855228 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr3 | 33628028 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os03g59070 |
| SBS | Chr3 | 36344507 | T-C | HOMO | INTRON | |
| SBS | Chr3 | 6581337 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g12460 |
| SBS | Chr3 | 6879711 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 9517651 | A-T | HOMO | INTRON | |
| SBS | Chr4 | 12044000 | A-G | HET | INTERGENIC | |
| SBS | Chr4 | 13091793 | T-C | HOMO | INTRON | |
| SBS | Chr4 | 14201602 | T-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 17698948 | T-A | HET | INTERGENIC | |
| SBS | Chr4 | 19462099 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 22697891 | C-A | HET | INTERGENIC | |
| SBS | Chr4 | 33507551 | G-T | HET | INTERGENIC | |
| SBS | Chr5 | 17833199 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 5139023 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 14072701 | C-A | HET | INTERGENIC | |
| SBS | Chr6 | 22424098 | T-G | HET | INTERGENIC | |
| SBS | Chr6 | 30713687 | G-T | HET | INTERGENIC | |
| SBS | Chr6 | 30713688 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 30713689 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 6824316 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os06g12590 |
| SBS | Chr6 | 6824319 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os06g12590 |
| SBS | Chr6 | 7316597 | C-A | HET | INTERGENIC | |
| SBS | Chr6 | 7316598 | T-G | HET | INTERGENIC | |
| SBS | Chr6 | 7316599 | A-T | HET | INTERGENIC | |
| SBS | Chr6 | 7429697 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 27547363 | C-A | HOMO | INTRON | |
| SBS | Chr7 | 4283338 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 5554353 | T-C | HET | INTRON | |
| SBS | Chr8 | 15471670 | C-G | HET | INTERGENIC | |
| SBS | Chr8 | 16929054 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr8 | 1806451 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 26303279 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 5057293 | T-A | HET | INTERGENIC | |
| SBS | Chr9 | 12265040 | T-A | HET | INTRON | |
| SBS | Chr9 | 13507587 | G-C | HOMO | NON_SYNONYMOUS_CODING | LOC_Os09g22350 |
| SBS | Chr9 | 13551576 | C-A | HET | INTERGENIC | |
| SBS | Chr9 | 17580736 | T-A | HET | INTERGENIC |
Deletions: 23
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr11 | 2354353 | 2354354 | 1 | |
| Deletion | Chr11 | 3001998 | 3001999 | 1 | |
| Deletion | Chr3 | 4141568 | 4141569 | 1 | |
| Deletion | Chr9 | 5223658 | 5223659 | 1 | |
| Deletion | Chr10 | 7028293 | 7028294 | 1 | |
| Deletion | Chr9 | 8501638 | 8501640 | 2 | |
| Deletion | Chr10 | 8773122 | 8773126 | 4 | LOC_Os10g17400 |
| Deletion | Chr11 | 10280807 | 10280817 | 10 | |
| Deletion | Chr3 | 11237637 | 11237657 | 20 | |
| Deletion | Chr12 | 12302108 | 12302122 | 14 | |
| Deletion | Chr9 | 14494966 | 14494967 | 1 | |
| Deletion | Chr7 | 18561182 | 18561183 | 1 | LOC_Os07g31340 |
| Deletion | Chr2 | 22735043 | 22735046 | 3 | |
| Deletion | Chr11 | 22812858 | 22812867 | 9 | |
| Deletion | Chr1 | 24891112 | 24891150 | 38 | |
| Deletion | Chr1 | 26513559 | 26513560 | 1 | |
| Deletion | Chr6 | 26584863 | 26584883 | 20 | |
| Deletion | Chr7 | 26757310 | 26757311 | 1 | |
| Deletion | Chr8 | 27970626 | 27970628 | 2 | |
| Deletion | Chr7 | 28438001 | 28445000 | 6999 | 2 |
| Deletion | Chr4 | 31250853 | 31250855 | 2 | LOC_Os04g52560 |
| Deletion | Chr3 | 34484716 | 34484721 | 5 | |
| Deletion | Chr2 | 35809575 | 35809580 | 5 |
Insertions: 6
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr10 | 8106863 | 8685880 | |
| Inversion | Chr11 | 16376707 | 17008514 |
Translocations: 4
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr7 | 11781006 | Chr1 | 20049378 | |
| Translocation | Chr11 | 15778951 | Chr9 | 11329510 | |
| Translocation | Chr12 | 15976550 | Chr7 | 11781189 | |
| Translocation | Chr12 | 15977128 | Chr7 | 11780997 |
