Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2658-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2658-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 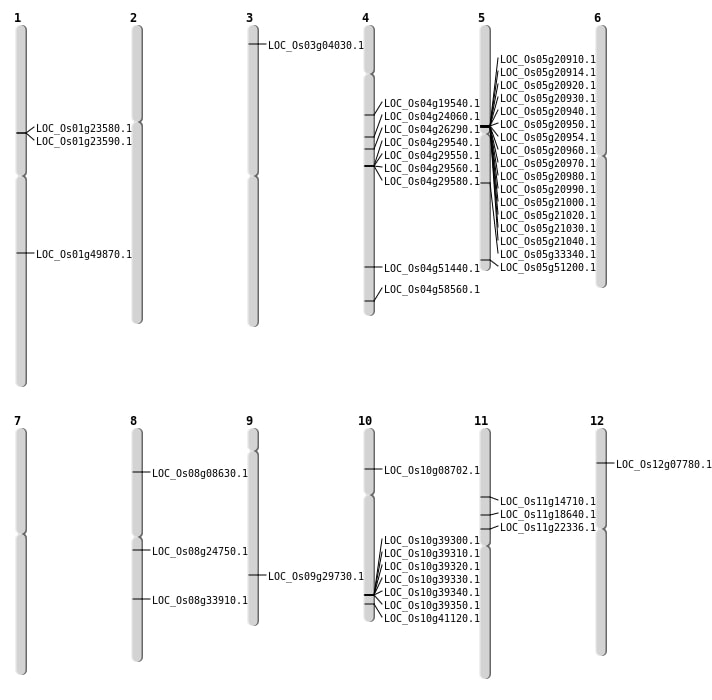
|
| Variant Information |
|---|
Single base substitutions: 53
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 10604210 | T-C | Homo | ||
| SBS | Chr1 | 12571703 | A-G | HET | ||
| SBS | Chr1 | 15968097 | T-A | Homo | ||
| SBS | Chr1 | 21476490 | A-T | HET | ||
| SBS | Chr1 | 28645403 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g49870 |
| SBS | Chr10 | 23012800 | G-T | HET | ||
| SBS | Chr10 | 23185776 | C-T | HET | ||
| SBS | Chr10 | 4720042 | A-C | HET | NON_SYNONYMOUS_CODING | LOC_Os10g08702 |
| SBS | Chr11 | 10522657 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g18640 |
| SBS | Chr11 | 14480752 | G-A | HET | ||
| SBS | Chr11 | 16417094 | G-T | HET | ||
| SBS | Chr11 | 23325086 | G-A | HET | ||
| SBS | Chr11 | 24660491 | G-A | HET | ||
| SBS | Chr11 | 8297218 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g14710 |
| SBS | Chr12 | 10225740 | T-G | HET | ||
| SBS | Chr12 | 23312383 | C-T | HET | ||
| SBS | Chr12 | 24136237 | A-G | HET | ||
| SBS | Chr12 | 3932656 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g07780 |
| SBS | Chr2 | 11385050 | C-T | Homo | ||
| SBS | Chr2 | 26024474 | A-T | Homo | ||
| SBS | Chr3 | 1838424 | C-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os03g04030 |
| SBS | Chr3 | 24207935 | G-A | HET | ||
| SBS | Chr3 | 31392958 | G-T | HET | ||
| SBS | Chr3 | 35945318 | G-T | HET | ||
| SBS | Chr4 | 14216517 | T-A | Homo | ||
| SBS | Chr4 | 15325223 | C-A | Homo | STOP_GAINED | LOC_Os04g26290 |
| SBS | Chr4 | 19045630 | C-T | Homo | ||
| SBS | Chr4 | 21745322 | T-A | HET | ||
| SBS | Chr4 | 25251352 | C-T | Homo | ||
| SBS | Chr4 | 30285421 | G-A | Homo | ||
| SBS | Chr4 | 8765173 | A-G | Homo | ||
| SBS | Chr5 | 12802298 | G-A | HET | ||
| SBS | Chr5 | 14444785 | G-A | Homo | ||
| SBS | Chr5 | 18217872 | T-C | Homo | ||
| SBS | Chr5 | 19556011 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g33340 |
| SBS | Chr5 | 20369899 | T-A | HET | ||
| SBS | Chr5 | 25749517 | C-T | HET | ||
| SBS | Chr5 | 29369399 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os05g51200 |
| SBS | Chr5 | 7118297 | T-C | HET | ||
| SBS | Chr6 | 20211010 | G-A | Homo | ||
| SBS | Chr6 | 20222556 | G-A | HET | ||
| SBS | Chr6 | 25864775 | G-A | HET | ||
| SBS | Chr7 | 15733876 | C-T | Homo | ||
| SBS | Chr7 | 66470 | T-A | HET | ||
| SBS | Chr8 | 17931019 | C-T | HET | ||
| SBS | Chr8 | 180658 | C-T | HET | ||
| SBS | Chr8 | 21219681 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os08g33910 |
| SBS | Chr8 | 23025642 | G-A | Homo | ||
| SBS | Chr8 | 4987631 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os08g08630 |
| SBS | Chr8 | 5927471 | G-A | HET | ||
| SBS | Chr9 | 4155116 | C-T | Homo | ||
| SBS | Chr9 | 5191377 | C-A | HET | ||
| SBS | Chr9 | 8587553 | T-C | HET |
Deletions: 23
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr12 | 1126153 | 1126156 | 3 | |
| Deletion | Chr9 | 1889397 | 1889398 | 1 | |
| Deletion | Chr6 | 5394300 | 5394317 | 17 | |
| Deletion | Chr6 | 5731403 | 5731419 | 16 | |
| Deletion | Chr1 | 10604406 | 10604414 | 8 | |
| Deletion | Chr5 | 12279001 | 12360000 | 80999 | 12 |
| Deletion | Chr5 | 12369001 | 12380000 | 10999 | 3 |
| Deletion | Chr11 | 12805330 | 12805332 | 2 | LOC_Os11g22336 |
| Deletion | Chr1 | 13246001 | 13262000 | 15999 | 2 |
| Deletion | Chr4 | 13745766 | 13745772 | 6 | LOC_Os04g24060 |
| Deletion | Chr6 | 14606073 | 14606090 | 17 | |
| Deletion | Chr8 | 14976451 | 14976457 | 6 | LOC_Os08g24750 |
| Deletion | Chr9 | 16231304 | 16231305 | 1 | |
| Deletion | Chr8 | 17299048 | 17299138 | 90 | |
| Deletion | Chr4 | 17552001 | 17587000 | 34999 | 4 |
| Deletion | Chr9 | 18081127 | 18081132 | 5 | LOC_Os09g29730 |
| Deletion | Chr3 | 18897930 | 18897932 | 2 | |
| Deletion | Chr12 | 19263661 | 19263673 | 12 | |
| Deletion | Chr10 | 20971001 | 20998000 | 26999 | 6 |
| Deletion | Chr10 | 22086847 | 22086851 | 4 | LOC_Os10g41120 |
| Deletion | Chr5 | 29007594 | 29007603 | 9 | |
| Deletion | Chr4 | 30464174 | 30464189 | 15 | LOC_Os04g51440 |
| Deletion | Chr3 | 34983321 | 34983323 | 2 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr10 | 20447277 | 20447300 | 24 | |
| Insertion | Chr12 | 1632927 | 1632932 | 6 | |
| Insertion | Chr12 | 228150 | 228151 | 2 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr4 | 17552436 | 17644272 | |
| Inversion | Chr4 | 17586683 | 17644273 | LOC_Os04g29580 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 10750969 | Chr4 | 34819821 | 2 |
| Translocation | Chr8 | 27565430 | Chr4 | 10877213 | 2 |
| Translocation | Chr8 | 27565450 | Chr4 | 10875680 | 2 |
