Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2659-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2659-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 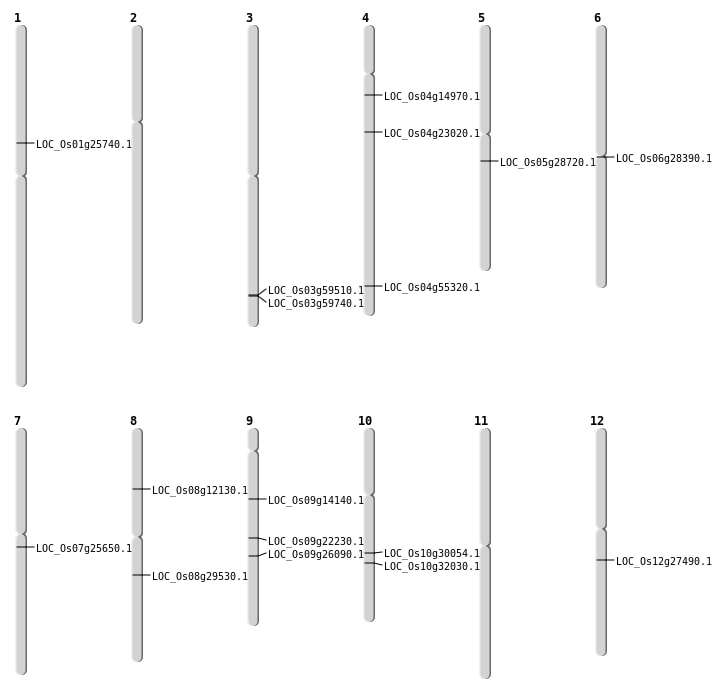
|
| Variant Information |
|---|
Single base substitutions: 62
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 23217311 | G-A | Homo | ||
| SBS | Chr10 | 21306514 | C-A | HET | ||
| SBS | Chr10 | 9235004 | C-T | HET | ||
| SBS | Chr11 | 14852190 | C-T | HET | ||
| SBS | Chr12 | 10862208 | C-A | HET | ||
| SBS | Chr12 | 12166233 | T-G | HET | ||
| SBS | Chr12 | 12166235 | A-C | HET | ||
| SBS | Chr12 | 12520176 | A-G | HET | ||
| SBS | Chr12 | 15495816 | A-C | HET | ||
| SBS | Chr12 | 23121146 | T-A | HET | ||
| SBS | Chr12 | 654640 | C-A | HET | ||
| SBS | Chr12 | 7862766 | C-T | HET | ||
| SBS | Chr12 | 8783949 | A-G | HET | ||
| SBS | Chr12 | 9681578 | C-T | HET | ||
| SBS | Chr2 | 12584953 | T-G | Homo | ||
| SBS | Chr2 | 14119805 | G-A | HET | ||
| SBS | Chr2 | 17267034 | G-C | HET | ||
| SBS | Chr2 | 17464852 | A-G | Homo | ||
| SBS | Chr2 | 20529303 | G-A | HET | ||
| SBS | Chr2 | 23473348 | C-T | HET | ||
| SBS | Chr2 | 30823133 | C-A | HET | ||
| SBS | Chr2 | 30823134 | G-A | HET | ||
| SBS | Chr3 | 10370593 | C-A | HET | ||
| SBS | Chr3 | 11658533 | A-T | HET | ||
| SBS | Chr3 | 12436286 | C-T | Homo | ||
| SBS | Chr3 | 17376767 | A-G | HET | ||
| SBS | Chr3 | 18064005 | A-T | HET | ||
| SBS | Chr3 | 18064007 | T-G | HET | ||
| SBS | Chr3 | 2408863 | G-T | HET | ||
| SBS | Chr3 | 24941984 | G-A | Homo | ||
| SBS | Chr3 | 24941985 | C-T | Homo | ||
| SBS | Chr3 | 29025841 | C-T | Homo | ||
| SBS | Chr3 | 31940659 | C-T | HET | ||
| SBS | Chr3 | 33871859 | G-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os03g59510 |
| SBS | Chr4 | 13067954 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g23020 |
| SBS | Chr4 | 21109563 | C-T | HET | ||
| SBS | Chr4 | 29856679 | C-T | HET | ||
| SBS | Chr4 | 7068837 | A-C | HET | ||
| SBS | Chr4 | 7424000 | T-C | HET | ||
| SBS | Chr5 | 16846883 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os05g28720 |
| SBS | Chr5 | 24853505 | T-G | HET | ||
| SBS | Chr5 | 4179874 | T-C | HET | ||
| SBS | Chr5 | 4191452 | T-G | HET | ||
| SBS | Chr5 | 5907370 | G-A | HET | ||
| SBS | Chr6 | 14280966 | T-A | HET | ||
| SBS | Chr6 | 14729607 | G-T | HET | ||
| SBS | Chr6 | 21510611 | G-A | HET | ||
| SBS | Chr6 | 21658664 | T-A | HET | ||
| SBS | Chr6 | 26788694 | A-G | HET | ||
| SBS | Chr7 | 14708040 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os07g25650 |
| SBS | Chr7 | 28506560 | G-A | HET | ||
| SBS | Chr7 | 5789409 | T-C | HET | ||
| SBS | Chr8 | 17662898 | G-T | HET | ||
| SBS | Chr8 | 18105086 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os08g29530 |
| SBS | Chr8 | 19143032 | T-A | HET | ||
| SBS | Chr8 | 22463919 | G-T | Homo | ||
| SBS | Chr9 | 10259094 | C-T | HET | ||
| SBS | Chr9 | 15693204 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os09g26090 |
| SBS | Chr9 | 22634894 | G-A | HET | ||
| SBS | Chr9 | 7984942 | C-T | HET | ||
| SBS | Chr9 | 8376866 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os09g14140 |
| SBS | Chr9 | 8682605 | A-G | HET |
Deletions: 14
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr1 | 796632 | 796634 | 2 | |
| Deletion | Chr4 | 919745 | 919757 | 12 | |
| Deletion | Chr7 | 2847030 | 2847031 | 1 | |
| Deletion | Chr4 | 9088817 | 9088819 | 2 | |
| Deletion | Chr8 | 10721454 | 10721455 | 1 | |
| Deletion | Chr10 | 16831264 | 16831276 | 12 | LOC_Os10g32030 |
| Deletion | Chr7 | 22054665 | 22054667 | 2 | |
| Deletion | Chr11 | 23109845 | 23109846 | 1 | |
| Deletion | Chr3 | 24941870 | 24941872 | 2 | |
| Deletion | Chr3 | 24948172 | 24948176 | 4 | |
| Deletion | Chr2 | 27957220 | 27957224 | 4 | |
| Deletion | Chr6 | 29719276 | 29719280 | 4 | |
| Deletion | Chr3 | 34009001 | 34022000 | 12999 | LOC_Os03g59740 |
| Deletion | Chr3 | 34981365 | 34981366 | 1 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr4 | 32929666 | 32929666 | 1 | |
| Insertion | Chr4 | 8409851 | 8409851 | 1 | LOC_Os04g14970 |
| Insertion | Chr5 | 10007414 | 10007414 | 1 | |
| Insertion | Chr6 | 25354194 | 25354195 | 2 |
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr4 | 5150782 | 5497977 | |
| Inversion | Chr4 | 5150958 | 5497987 | |
| Inversion | Chr10 | 15609345 | 15754220 | LOC_Os10g30054 |
| Inversion | Chr10 | 15609350 | 15754220 | LOC_Os10g30054 |
Translocations: 9
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr4 | 5150798 | Chr3 | 13830539 | |
| Translocation | Chr4 | 5150942 | Chr3 | 13830547 | |
| Translocation | Chr9 | 13448251 | Chr8 | 7145722 | 2 |
| Translocation | Chr10 | 13650187 | Chr6 | 16152481 | 2 |
| Translocation | Chr5 | 15913918 | Chr3 | 1302376 | |
| Translocation | Chr12 | 16182382 | Chr4 | 32905475 | 2 |
| Translocation | Chr4 | 17940237 | Chr2 | 13401460 | LOC_Os04g30040 |
| Translocation | Chr9 | 21246712 | Chr2 | 18115022 | |
| Translocation | Chr7 | 26695193 | Chr1 | 14588958 | 2 |
