Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2660-S-1 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2660-S-1 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 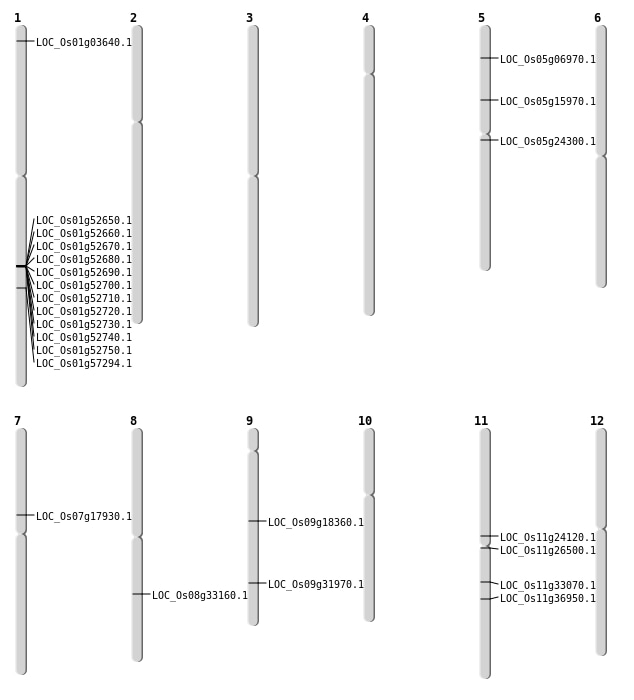
|
| Variant Information |
|---|
Single base substitutions: 47
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 19027873 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 19737218 | T-C | HET | INTERGENIC | |
| SBS | Chr1 | 28068264 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 29329544 | T-C | HET | INTRON | |
| SBS | Chr1 | 32063713 | T-A | HOMO | INTERGENIC | |
| SBS | Chr10 | 8726525 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 21798356 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g36950 |
| SBS | Chr11 | 24774111 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 10915341 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 14430760 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 14948404 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 15043437 | C-G | HET | INTERGENIC | |
| SBS | Chr12 | 248452 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 18885656 | G-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 13326217 | C-A | HOMO | INTRON | |
| SBS | Chr3 | 16849124 | G-T | HET | INTRON | |
| SBS | Chr3 | 23835002 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 28369561 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 33256877 | G-A | HET | INTRON | |
| SBS | Chr4 | 12638042 | T-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 1367334 | C-A | HET | INTRON | |
| SBS | Chr4 | 19428291 | C-G | HOMO | INTERGENIC | |
| SBS | Chr4 | 7420905 | C-T | HOMO | INTRON | |
| SBS | Chr5 | 14062997 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g24300 |
| SBS | Chr5 | 16823315 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 24963282 | T-C | HET | INTERGENIC | |
| SBS | Chr5 | 24963283 | A-T | HET | INTERGENIC | |
| SBS | Chr5 | 9019343 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os05g15970 |
| SBS | Chr6 | 15262705 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 10608441 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os07g17930 |
| SBS | Chr7 | 18909743 | C-A | HET | INTERGENIC | |
| SBS | Chr7 | 4486523 | T-C | HET | INTRON | |
| SBS | Chr7 | 4486524 | C-T | HET | INTRON | |
| SBS | Chr7 | 8139444 | T-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr8 | 12696462 | T-C | HOMO | INTERGENIC | |
| SBS | Chr8 | 14518896 | G-T | HET | INTERGENIC | |
| SBS | Chr8 | 19492637 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr8 | 20626123 | T-G | HET | NON_SYNONYMOUS_CODING | LOC_Os08g33160 |
| SBS | Chr8 | 22564733 | G-T | HET | INTERGENIC | |
| SBS | Chr8 | 25750222 | T-C | HET | INTRON | |
| SBS | Chr8 | 26581080 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 11260000 | T-C | HOMO | NON_SYNONYMOUS_CODING | LOC_Os09g18360 |
| SBS | Chr9 | 11260005 | C-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os09g18360 |
| SBS | Chr9 | 14621063 | G-A | HOMO | INTERGENIC | |
| SBS | Chr9 | 19074656 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os09g31970 |
| SBS | Chr9 | 6591940 | G-A | HOMO | INTERGENIC | |
| SBS | Chr9 | 6591942 | A-T | HOMO | INTERGENIC |
Deletions: 17
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr1 | 507903 | 507904 | 1 | |
| Deletion | Chr5 | 562189 | 562193 | 4 | |
| Deletion | Chr2 | 2491818 | 2491823 | 5 | |
| Deletion | Chr10 | 3606127 | 3606134 | 7 | |
| Deletion | Chr5 | 3648026 | 3648040 | 14 | LOC_Os05g06970 |
| Deletion | Chr5 | 4409638 | 4409639 | 1 | |
| Deletion | Chr5 | 4492760 | 4492770 | 10 | |
| Deletion | Chr9 | 5755891 | 5755892 | 1 | |
| Deletion | Chr10 | 9215489 | 9215496 | 7 | |
| Deletion | Chr11 | 15191495 | 15191496 | 1 | LOC_Os11g26500 |
| Deletion | Chr8 | 17922675 | 17922699 | 24 | |
| Deletion | Chr11 | 19552343 | 19552344 | 1 | LOC_Os11g33070 |
| Deletion | Chr4 | 26431561 | 26431563 | 2 | |
| Deletion | Chr12 | 26860186 | 26860187 | 1 | |
| Deletion | Chr1 | 30257001 | 30349000 | 91999 | 12 |
| Deletion | Chr2 | 34850036 | 34850038 | 2 | |
| Deletion | Chr3 | 35090458 | 35090461 | 3 |
Insertions: 25
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr4 | 11372582 | 11372758 |
Translocations: 9
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr2 | 2071700 | Chr1 | 1500226 | LOC_Os01g03640 |
| Translocation | Chr10 | 3669339 | Chr4 | 33407149 | |
| Translocation | Chr2 | 7954238 | Chr1 | 15637503 | |
| Translocation | Chr7 | 10136799 | Chr2 | 30553950 | |
| Translocation | Chr7 | 10140869 | Chr2 | 30553874 | |
| Translocation | Chr12 | 13410062 | Chr11 | 19402557 | |
| Translocation | Chr11 | 13708389 | Chr6 | 11097874 | LOC_Os11g24120 |
| Translocation | Chr11 | 27437891 | Chr7 | 25323788 | |
| Translocation | Chr3 | 30923708 | Chr1 | 22551039 |
