Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2661-S-1 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2661-S-1 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 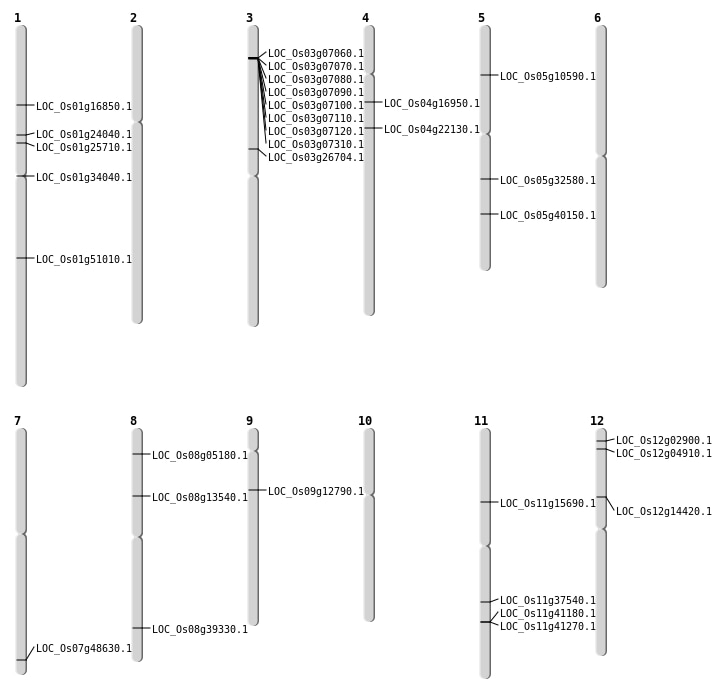
|
| Variant Information |
|---|
Single base substitutions: 38
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 13545895 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os01g24040 |
| SBS | Chr1 | 29310460 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g51010 |
| SBS | Chr1 | 37183017 | G-T | HOMO | INTRON | |
| SBS | Chr10 | 13679096 | C-A | HET | INTRON | |
| SBS | Chr10 | 22854623 | C-A | HET | INTERGENIC | |
| SBS | Chr10 | 4202020 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 7652730 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 9435241 | A-T | HET | INTERGENIC | |
| SBS | Chr11 | 10234392 | G-T | HET | INTERGENIC | |
| SBS | Chr11 | 16402640 | A-T | HET | INTERGENIC | |
| SBS | Chr11 | 20168091 | A-T | HET | INTERGENIC | |
| SBS | Chr11 | 23002125 | T-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 3618149 | T-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr11 | 8635998 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 8783850 | A-C | HET | INTERGENIC | |
| SBS | Chr12 | 11519557 | A-C | HET | INTERGENIC | |
| SBS | Chr12 | 1201890 | A-C | HOMO | UTR_5_PRIME | |
| SBS | Chr12 | 23787979 | T-A | HET | INTERGENIC | |
| SBS | Chr12 | 3540968 | T-C | HOMO | INTERGENIC | |
| SBS | Chr12 | 7473769 | G-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 29523171 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 33285713 | T-G | HET | INTERGENIC | |
| SBS | Chr4 | 17637119 | C-T | HET | INTRON | |
| SBS | Chr4 | 1836432 | T-C | HOMO | INTRON | |
| SBS | Chr4 | 26395625 | T-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 19075320 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g32580 |
| SBS | Chr5 | 26695763 | A-G | HET | INTRON | |
| SBS | Chr5 | 29886601 | T-A | HOMO | INTRON | |
| SBS | Chr5 | 7724375 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 16753820 | A-G | HOMO | INTERGENIC | |
| SBS | Chr7 | 14156455 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 17315477 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 18802709 | G-T | HET | INTRON | |
| SBS | Chr8 | 26016603 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 8049933 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os08g13540 |
| SBS | Chr9 | 18589761 | G-T | HET | INTERGENIC | |
| SBS | Chr9 | 18589762 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 3493491 | A-T | HET | INTERGENIC |
Deletions: 36
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr9 | 575583 | 575585 | 2 | |
| Deletion | Chr12 | 1063917 | 1063937 | 20 | LOC_Os12g02900 |
| Deletion | Chr4 | 1226852 | 1226858 | 6 | |
| Deletion | Chr8 | 2052119 | 2052120 | 1 | |
| Deletion | Chr8 | 2705801 | 2705804 | 3 | LOC_Os08g05180 |
| Deletion | Chr12 | 2978714 | 2978722 | 8 | |
| Deletion | Chr3 | 3595001 | 3641000 | 45999 | 8 |
| Deletion | Chr2 | 4601499 | 4601506 | 7 | |
| Deletion | Chr10 | 7671058 | 7671061 | 3 | |
| Deletion | Chr9 | 8061542 | 8061543 | 1 | |
| Deletion | Chr7 | 8275735 | 8275736 | 1 | |
| Deletion | Chr11 | 8893815 | 8893863 | 48 | LOC_Os11g15690 |
| Deletion | Chr6 | 8926385 | 8926386 | 1 | |
| Deletion | Chr4 | 9283415 | 9283416 | 1 | LOC_Os04g16950 |
| Deletion | Chr1 | 9622014 | 9622021 | 7 | LOC_Os01g16850 |
| Deletion | Chr5 | 9951669 | 9951671 | 2 | |
| Deletion | Chr8 | 12514536 | 12514537 | 1 | |
| Deletion | Chr2 | 12916297 | 12916303 | 6 | |
| Deletion | Chr11 | 13791019 | 13791036 | 17 | |
| Deletion | Chr9 | 13792878 | 13792880 | 2 | |
| Deletion | Chr5 | 13812545 | 13812580 | 35 | |
| Deletion | Chr1 | 14574548 | 14574550 | 2 | LOC_Os01g25710 |
| Deletion | Chr3 | 15524152 | 15524154 | 2 | |
| Deletion | Chr11 | 17500043 | 17500046 | 3 | |
| Deletion | Chr5 | 17562409 | 17562439 | 30 | |
| Deletion | Chr4 | 18132441 | 18132442 | 1 | |
| Deletion | Chr3 | 18947478 | 18947481 | 3 | |
| Deletion | Chr6 | 20927480 | 20927484 | 4 | |
| Deletion | Chr8 | 22576556 | 22576557 | 1 | |
| Deletion | Chr11 | 24686500 | 24686502 | 2 | LOC_Os11g41180 |
| Deletion | Chr11 | 24740711 | 24740737 | 26 | LOC_Os11g41270 |
| Deletion | Chr8 | 24855057 | 24855071 | 14 | LOC_Os08g39330 |
| Deletion | Chr8 | 25236241 | 25236242 | 1 | |
| Deletion | Chr12 | 25734842 | 25734843 | 1 | |
| Deletion | Chr8 | 26105604 | 26105605 | 1 | |
| Deletion | Chr1 | 36116743 | 36116744 | 1 |
Insertions: 24
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 31019728 | 31019728 | 1 | |
| Insertion | Chr1 | 871582 | 871591 | 10 | |
| Insertion | Chr10 | 15537597 | 15537597 | 1 | |
| Insertion | Chr10 | 2239293 | 2239340 | 48 | |
| Insertion | Chr10 | 5144804 | 5144805 | 2 | |
| Insertion | Chr11 | 13327713 | 13327713 | 1 | |
| Insertion | Chr11 | 3058725 | 3058725 | 1 | |
| Insertion | Chr12 | 10729497 | 10729527 | 31 | |
| Insertion | Chr12 | 18215775 | 18215776 | 2 | |
| Insertion | Chr2 | 16495023 | 16495023 | 1 | |
| Insertion | Chr2 | 20207445 | 20207445 | 1 | |
| Insertion | Chr2 | 23475478 | 23475478 | 1 | |
| Insertion | Chr2 | 34738868 | 34738883 | 16 | |
| Insertion | Chr3 | 22759606 | 22759617 | 12 | |
| Insertion | Chr4 | 11974491 | 11974491 | 1 | |
| Insertion | Chr4 | 17622261 | 17622308 | 48 | |
| Insertion | Chr4 | 8749036 | 8749036 | 1 | |
| Insertion | Chr4 | 9997072 | 9997111 | 40 | |
| Insertion | Chr5 | 29932065 | 29932065 | 1 | |
| Insertion | Chr5 | 9941426 | 9941426 | 1 | |
| Insertion | Chr6 | 19903339 | 19903348 | 10 | |
| Insertion | Chr6 | 21385147 | 21385147 | 1 | |
| Insertion | Chr7 | 7882486 | 7882486 | 1 | |
| Insertion | Chr9 | 7340127 | 7340129 | 3 | LOC_Os09g12790 |
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr5 | 6278539 | 6462357 | |
| Inversion | Chr5 | 6278560 | 6462360 | |
| Inversion | Chr4 | 32240786 | 32939701 | |
| Inversion | Chr4 | 32240797 | 32939713 |
Translocations: 25
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 2109751 | Chr9 | 9468878 | LOC_Os12g04910 |
| Translocation | Chr9 | 6382652 | Chr7 | 6237696 | |
| Translocation | Chr12 | 8221232 | Chr4 | 3485782 | LOC_Os12g14420 |
| Translocation | Chr8 | 8505944 | Chr3 | 2897018 | |
| Translocation | Chr12 | 10482998 | Chr7 | 29118623 | LOC_Os07g48630 |
| Translocation | Chr4 | 13171073 | Chr1 | 8783373 | |
| Translocation | Chr12 | 13373658 | Chr4 | 12545953 | LOC_Os04g22130 |
| Translocation | Chr6 | 13713679 | Chr2 | 2393042 | |
| Translocation | Chr8 | 14577417 | Chr6 | 8968045 | |
| Translocation | Chr8 | 15765978 | Chr3 | 15248058 | LOC_Os03g26704 |
| Translocation | Chr8 | 18074315 | Chr1 | 18737653 | LOC_Os01g34040 |
| Translocation | Chr6 | 18279856 | Chr4 | 8850351 | |
| Translocation | Chr12 | 19105199 | Chr3 | 14531802 | |
| Translocation | Chr8 | 20135723 | Chr1 | 2999832 | |
| Translocation | Chr12 | 20502586 | Chr8 | 1952438 | |
| Translocation | Chr11 | 22159796 | Chr8 | 10137906 | LOC_Os11g37540 |
| Translocation | Chr11 | 22532797 | Chr5 | 5767258 | LOC_Os05g10590 |
| Translocation | Chr7 | 23277372 | Chr3 | 13642981 | |
| Translocation | Chr12 | 23803678 | Chr3 | 22077710 | |
| Translocation | Chr4 | 26106170 | Chr1 | 10836412 | |
| Translocation | Chr4 | 27007922 | Chr2 | 28383914 | |
| Translocation | Chr12 | 27039704 | Chr2 | 7526584 | |
| Translocation | Chr11 | 27688464 | Chr5 | 23584250 | LOC_Os05g40150 |
| Translocation | Chr11 | 27757035 | Chr4 | 3131208 | |
| Translocation | Chr6 | 28538073 | Chr3 | 15949972 |
