Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2666-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2666-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 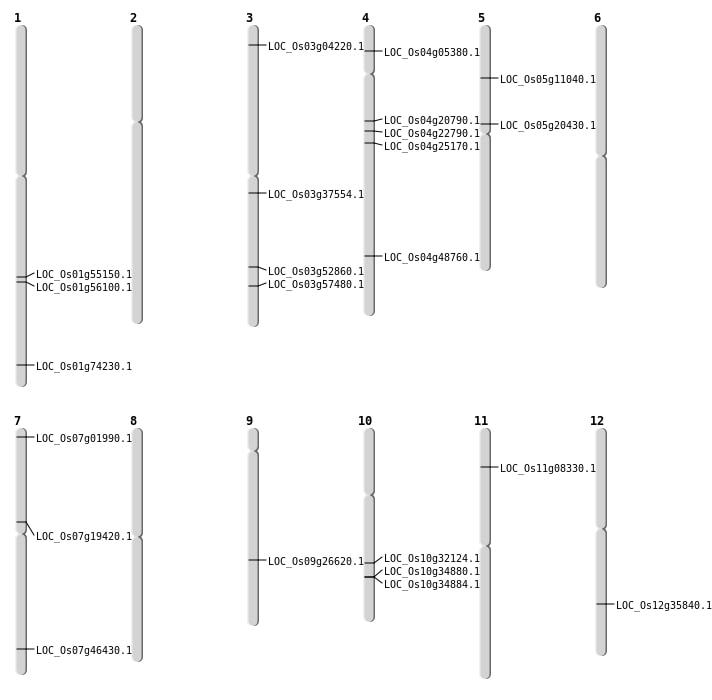
|
| Variant Information |
|---|
Single base substitutions: 98
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 18421985 | T-A | HET | ||
| SBS | Chr1 | 1891943 | C-T | HET | ||
| SBS | Chr1 | 20153958 | A-T | HET | ||
| SBS | Chr1 | 20697806 | A-T | HET | ||
| SBS | Chr1 | 20697806 | A-T | HET | INTERGENIC | |
| SBS | Chr1 | 23159960 | T-G | HET | ||
| SBS | Chr1 | 23159960 | T-G | HET | INTERGENIC | |
| SBS | Chr1 | 28575834 | C-T | HET | ||
| SBS | Chr1 | 32138485 | C-T | HET | ||
| SBS | Chr1 | 32138485 | C-T | HET | INTRON | |
| SBS | Chr1 | 37566701 | A-G | HET | ||
| SBS | Chr10 | 13053586 | C-T | HET | ||
| SBS | Chr10 | 13203684 | G-A | HET | ||
| SBS | Chr10 | 13203684 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr10 | 20419798 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 5040174 | C-A | HET | ||
| SBS | Chr10 | 5040174 | C-A | HET | INTERGENIC | |
| SBS | Chr10 | 8867827 | C-A | HET | ||
| SBS | Chr10 | 8867827 | C-A | HET | INTERGENIC | |
| SBS | Chr11 | 15215036 | C-G | HET | ||
| SBS | Chr11 | 20906425 | A-T | HET | ||
| SBS | Chr11 | 20906425 | A-T | HET | INTERGENIC | |
| SBS | Chr11 | 4394509 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os11g08330 |
| SBS | Chr12 | 21841571 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g35840 |
| SBS | Chr12 | 4215861 | A-G | HET | ||
| SBS | Chr12 | 4215861 | A-G | HET | INTRON | |
| SBS | Chr12 | 4719648 | C-A | HET | ||
| SBS | Chr12 | 4719648 | C-A | HET | INTERGENIC | |
| SBS | Chr2 | 20212545 | G-A | HET | ||
| SBS | Chr2 | 20212545 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 20696147 | G-A | HET | ||
| SBS | Chr2 | 20696147 | G-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 2890681 | G-T | Homo | ||
| SBS | Chr2 | 2890681 | G-T | HOMO | INTRON | |
| SBS | Chr3 | 13966957 | C-T | HET | ||
| SBS | Chr3 | 13966957 | C-T | HET | INTRON | |
| SBS | Chr3 | 21987387 | G-A | HET | ||
| SBS | Chr3 | 23651742 | G-A | HET | ||
| SBS | Chr3 | 24811687 | T-G | HET | ||
| SBS | Chr3 | 24811687 | T-G | HET | INTRON | |
| SBS | Chr3 | 32771600 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os03g57480 |
| SBS | Chr4 | 12905238 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g22790 |
| SBS | Chr4 | 13439346 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr4 | 14548274 | T-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os04g25170 |
| SBS | Chr4 | 14548274 | T-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os04g25170 |
| SBS | Chr4 | 21127408 | T-A | HET | ||
| SBS | Chr4 | 21127408 | T-A | HET | INTERGENIC | |
| SBS | Chr4 | 24463387 | G-A | Homo | ||
| SBS | Chr4 | 24463387 | G-A | HOMO | INTRON | |
| SBS | Chr4 | 29084546 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g48760 |
| SBS | Chr4 | 30612116 | G-A | HET | ||
| SBS | Chr4 | 31805012 | C-T | HET | ||
| SBS | Chr4 | 33613056 | C-T | HET | ||
| SBS | Chr4 | 4508022 | A-T | HET | ||
| SBS | Chr4 | 5015787 | C-A | HET | ||
| SBS | Chr4 | 5015787 | C-A | HET | INTERGENIC | |
| SBS | Chr5 | 10166310 | A-T | Homo | ||
| SBS | Chr5 | 10166310 | A-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 11988951 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os05g20430 |
| SBS | Chr5 | 11988951 | G-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os05g20430 |
| SBS | Chr5 | 5559279 | T-G | HET | ||
| SBS | Chr5 | 5559279 | T-G | HET | INTERGENIC | |
| SBS | Chr5 | 6189526 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g11040 |
| SBS | Chr6 | 24721282 | T-A | Homo | ||
| SBS | Chr6 | 24721282 | T-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 4048626 | C-T | HET | ||
| SBS | Chr6 | 4048626 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 8495887 | G-T | HET | ||
| SBS | Chr6 | 9526565 | G-T | HET | ||
| SBS | Chr7 | 10312621 | T-A | Homo | ||
| SBS | Chr7 | 10312621 | T-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 10312622 | G-A | Homo | ||
| SBS | Chr7 | 10312622 | G-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 10480702 | C-T | HET | ||
| SBS | Chr7 | 11489776 | G-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os07g19420 |
| SBS | Chr7 | 22094637 | C-A | HET | ||
| SBS | Chr7 | 22094637 | C-A | HET | INTERGENIC | |
| SBS | Chr7 | 2458484 | T-G | Homo | ||
| SBS | Chr7 | 3231286 | C-T | HET | ||
| SBS | Chr7 | 3231286 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr7 | 3366552 | A-T | Homo | ||
| SBS | Chr7 | 3366552 | A-T | HOMO | INTRON | |
| SBS | Chr8 | 10904192 | C-T | HET | INTRON | |
| SBS | Chr8 | 15964272 | C-G | HET | ||
| SBS | Chr8 | 15964272 | C-G | HET | INTERGENIC | |
| SBS | Chr8 | 2317753 | T-A | HET | ||
| SBS | Chr8 | 2317753 | T-A | HET | INTERGENIC | |
| SBS | Chr8 | 24089478 | C-T | HET | ||
| SBS | Chr9 | 16153647 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os09g26620 |
| SBS | Chr9 | 2270314 | C-A | HET | ||
| SBS | Chr9 | 2270314 | C-A | HET | INTRON | |
| SBS | Chr9 | 4621560 | A-G | HET | ||
| SBS | Chr9 | 641106 | C-T | HET | ||
| SBS | Chr9 | 641106 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 644892 | A-G | HET | ||
| SBS | Chr9 | 644892 | A-G | HET | INTERGENIC | |
| SBS | Chr9 | 9676857 | G-T | Homo | ||
| SBS | Chr9 | 9676857 | G-T | HOMO | INTERGENIC |
Deletions: 45
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr7 | 605204 | 605212 | 8 | 2 |
| Deletion | Chr5 | 1771724 | 1771744 | 20 | |
| Deletion | Chr3 | 1939037 | 1939058 | 21 | LOC_Os03g04220 |
| Deletion | Chr4 | 2692630 | 2692634 | 4 | LOC_Os04g05380 |
| Deletion | Chr3 | 3699591 | 3699597 | 6 | 2 |
| Deletion | Chr8 | 5095144 | 5095145 | 1 | 2 |
| Deletion | Chr9 | 7316960 | 7316961 | 1 | |
| Deletion | Chr2 | 7956138 | 7956139 | 1 | 2 |
| Deletion | Chr8 | 8686659 | 8686660 | 1 | 2 |
| Deletion | Chr5 | 9434136 | 9434144 | 8 | 2 |
| Deletion | Chr4 | 10755647 | 10755654 | 7 | |
| Deletion | Chr3 | 10788258 | 10788264 | 6 | |
| Deletion | Chr4 | 11663448 | 11663451 | 3 | LOC_Os04g20790 |
| Deletion | Chr12 | 11800702 | 11800710 | 8 | 2 |
| Deletion | Chr7 | 11804591 | 11804600 | 9 | 2 |
| Deletion | Chr1 | 12695248 | 12695254 | 6 | |
| Deletion | Chr10 | 14155163 | 14155164 | 1 | |
| Deletion | Chr1 | 16175671 | 16175672 | 1 | |
| Deletion | Chr4 | 16656096 | 16656100 | 4 | |
| Deletion | Chr1 | 17325886 | 17325887 | 1 | 2 |
| Deletion | Chr11 | 18074281 | 18074283 | 2 | 2 |
| Deletion | Chr10 | 18607001 | 18620000 | 12999 | 2 |
| Deletion | Chr1 | 18701437 | 18701440 | 3 | 2 |
| Deletion | Chr12 | 18860366 | 18860377 | 11 | |
| Deletion | Chr10 | 19015843 | 19015849 | 6 | |
| Deletion | Chr1 | 19761982 | 19761985 | 3 | 2 |
| Deletion | Chr1 | 20682259 | 20682265 | 6 | |
| Deletion | Chr1 | 20737313 | 20737316 | 3 | 2 |
| Deletion | Chr3 | 20834733 | 20834734 | 1 | |
| Deletion | Chr5 | 21041230 | 21041247 | 17 | 2 |
| Deletion | Chr3 | 21416922 | 21416923 | 1 | |
| Deletion | Chr6 | 24354331 | 24354336 | 5 | 2 |
| Deletion | Chr6 | 24613289 | 24613290 | 1 | |
| Deletion | Chr3 | 25350185 | 25350189 | 4 | |
| Deletion | Chr6 | 25874329 | 25874338 | 9 | 2 |
| Deletion | Chr11 | 26470538 | 26470553 | 15 | |
| Deletion | Chr3 | 27112892 | 27112899 | 7 | 2 |
| Deletion | Chr7 | 27702804 | 27702806 | 2 | LOC_Os07g46430 |
| Deletion | Chr3 | 27849846 | 27849863 | 17 | |
| Deletion | Chr8 | 27883310 | 27883320 | 10 | |
| Deletion | Chr6 | 28214001 | 28226000 | 11999 | |
| Deletion | Chr1 | 28441023 | 28441034 | 11 | |
| Deletion | Chr3 | 30317157 | 30317177 | 20 | 2 |
| Deletion | Chr4 | 31847393 | 31847479 | 86 | |
| Deletion | Chr3 | 32522079 | 32522080 | 1 |
Insertions: 13
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 3945123 | 3945124 | 2 | 2 |
| Insertion | Chr1 | 3945123 | 3945124 | 2 | 2 |
| Insertion | Chr1 | 43000361 | 43000361 | 1 | LOC_Os01g74230 |
| Insertion | Chr12 | 426978 | 426982 | 5 | |
| Insertion | Chr2 | 24310552 | 24310552 | 1 | |
| Insertion | Chr3 | 22759606 | 22759617 | 12 | |
| Insertion | Chr4 | 1058037 | 1058037 | 1 | |
| Insertion | Chr4 | 3133414 | 3133416 | 3 | 2 |
| Insertion | Chr4 | 3133414 | 3133416 | 3 | 2 |
| Insertion | Chr8 | 4957394 | 4957399 | 6 | 2 |
| Insertion | Chr8 | 4957394 | 4957399 | 6 | 2 |
| Insertion | Chr9 | 13061771 | 13061772 | 2 | 2 |
| Insertion | Chr9 | 13061771 | 13061772 | 2 | 2 |
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr10 | 16879281 | 17459431 | 2 |
| Inversion | Chr3 | 20831261 | 21068189 | |
| Inversion | Chr3 | 20831273 | 21068192 | 2 |
| Inversion | Chr1 | 31730291 | 32301893 | LOC_Os01g55150 |
| Inversion | Chr1 | 31730294 | 32301907 | 2 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 6457667 | Chr10 | 3607567 | |
| Translocation | Chr11 | 9323569 | Chr6 | 23180839 |
