Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2667-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2667-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 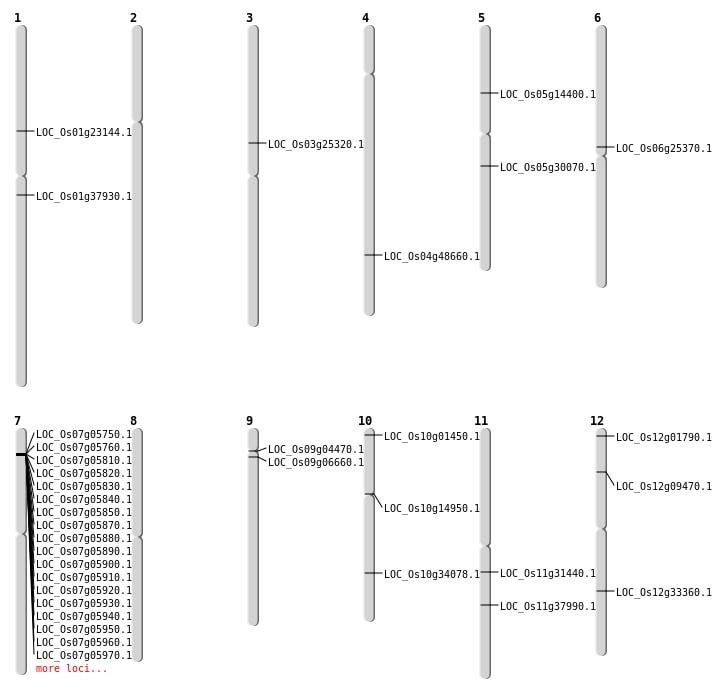
|
| Variant Information |
|---|
Single base substitutions: 108
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 11334190 | G-C | Homo | ||
| SBS | Chr1 | 11334190 | G-C | HOMO | INTRON | |
| SBS | Chr1 | 12040921 | T-C | Homo | ||
| SBS | Chr1 | 12040921 | T-C | HOMO | INTERGENIC | |
| SBS | Chr1 | 21249906 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g37930 |
| SBS | Chr1 | 22493925 | C-G | HET | ||
| SBS | Chr1 | 29497419 | A-G | Homo | ||
| SBS | Chr1 | 29497419 | A-G | HOMO | INTERGENIC | |
| SBS | Chr1 | 30580943 | G-T | HET | ||
| SBS | Chr1 | 32363817 | A-T | HET | ||
| SBS | Chr1 | 36324589 | G-C | HET | ||
| SBS | Chr1 | 953179 | G-A | HET | ||
| SBS | Chr10 | 12845922 | G-A | HET | ||
| SBS | Chr10 | 12845922 | G-A | HET | SPLICE_SITE_REGION | |
| SBS | Chr10 | 15317901 | T-C | HET | ||
| SBS | Chr10 | 15317901 | T-C | HET | INTRON | |
| SBS | Chr10 | 272497 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os10g01450 |
| SBS | Chr10 | 272497 | G-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os10g01450 |
| SBS | Chr11 | 15078255 | G-A | HET | ||
| SBS | Chr11 | 18180309 | G-T | HET | ||
| SBS | Chr11 | 20945524 | G-A | HET | INTRON | |
| SBS | Chr11 | 27811016 | T-A | HET | ||
| SBS | Chr12 | 12101219 | G-A | HET | ||
| SBS | Chr12 | 12101219 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 18071896 | G-C | HET | ||
| SBS | Chr12 | 18071896 | G-C | HET | INTERGENIC | |
| SBS | Chr12 | 4968337 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os12g09470 |
| SBS | Chr12 | 5013862 | G-A | HET | ||
| SBS | Chr12 | 5013862 | G-A | HET | UTR_3_PRIME | |
| SBS | Chr2 | 20353988 | A-G | HET | ||
| SBS | Chr2 | 20353988 | A-G | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 26720549 | C-T | HET | ||
| SBS | Chr2 | 26720549 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 30980440 | G-C | HET | ||
| SBS | Chr2 | 31974692 | C-T | HET | ||
| SBS | Chr2 | 31974692 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 32063413 | T-C | HET | ||
| SBS | Chr2 | 8443091 | G-T | HET | ||
| SBS | Chr3 | 11999278 | T-G | HET | ||
| SBS | Chr3 | 11999278 | T-G | HET | INTERGENIC | |
| SBS | Chr3 | 12451475 | T-A | HET | ||
| SBS | Chr3 | 12451475 | T-A | HET | INTERGENIC | |
| SBS | Chr3 | 13956805 | A-G | HET | ||
| SBS | Chr3 | 13956805 | A-G | HET | SYNONYMOUS_CODING | |
| SBS | Chr3 | 14454862 | T-G | HET | NON_SYNONYMOUS_CODING | LOC_Os03g25320 |
| SBS | Chr3 | 16391822 | T-A | HET | ||
| SBS | Chr3 | 16391822 | T-A | HET | INTERGENIC | |
| SBS | Chr3 | 16962158 | G-A | HET | ||
| SBS | Chr3 | 36195289 | G-A | HET | ||
| SBS | Chr3 | 9184654 | A-G | HET | ||
| SBS | Chr4 | 14851317 | G-A | Homo | ||
| SBS | Chr4 | 14851317 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 21776572 | G-A | Homo | ||
| SBS | Chr4 | 21776572 | G-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 22957084 | A-G | HET | ||
| SBS | Chr4 | 23874808 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 26393515 | C-A | HET | ||
| SBS | Chr4 | 26553387 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr4 | 27780665 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 28979118 | C-A | HET | ||
| SBS | Chr4 | 29013810 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g48660 |
| SBS | Chr4 | 4180235 | C-G | Homo | ||
| SBS | Chr4 | 4180235 | C-G | HOMO | INTERGENIC | |
| SBS | Chr4 | 4185433 | C-G | HOMO | UTR_3_PRIME | |
| SBS | Chr4 | 6174427 | G-T | HET | ||
| SBS | Chr5 | 17394550 | T-C | HOMO | NON_SYNONYMOUS_CODING | LOC_Os05g30070 |
| SBS | Chr5 | 17394550 | T-C | Homo | NON_SYNONYMOUS_CODING | LOC_Os05g30070 |
| SBS | Chr5 | 23414631 | A-C | Homo | ||
| SBS | Chr5 | 23414631 | A-C | HOMO | INTERGENIC | |
| SBS | Chr5 | 310454 | C-A | HET | ||
| SBS | Chr5 | 8159933 | T-C | Homo | ||
| SBS | Chr5 | 8159933 | T-C | HOMO | INTERGENIC | |
| SBS | Chr6 | 12977409 | C-A | HET | ||
| SBS | Chr6 | 13062251 | C-T | HET | ||
| SBS | Chr6 | 14860784 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os06g25370 |
| SBS | Chr6 | 14860784 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os06g25370 |
| SBS | Chr6 | 2396521 | G-A | HET | ||
| SBS | Chr6 | 2396521 | G-A | HET | UTR_3_PRIME | |
| SBS | Chr6 | 25627351 | A-G | HET | ||
| SBS | Chr6 | 31136604 | A-G | Homo | ||
| SBS | Chr6 | 31136604 | A-G | HOMO | INTERGENIC | |
| SBS | Chr6 | 6834121 | C-T | Homo | ||
| SBS | Chr6 | 6834121 | C-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 8351093 | A-T | Homo | ||
| SBS | Chr6 | 8351093 | A-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 10570155 | T-C | HET | ||
| SBS | Chr7 | 12886092 | C-G | HET | INTERGENIC | |
| SBS | Chr7 | 15372330 | G-A | HET | ||
| SBS | Chr7 | 15372330 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 160671 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 2790773 | A-C | HET | INTERGENIC | |
| SBS | Chr7 | 3592723 | T-C | HET | ||
| SBS | Chr7 | 3592723 | T-C | HET | INTRON | |
| SBS | Chr7 | 6676403 | G-A | HET | ||
| SBS | Chr7 | 6676403 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 9835937 | C-T | HET | ||
| SBS | Chr8 | 13697075 | T-A | HET | ||
| SBS | Chr8 | 21596089 | T-A | HET | ||
| SBS | Chr8 | 21596089 | T-A | HET | INTERGENIC | |
| SBS | Chr8 | 24746569 | C-A | HET | ||
| SBS | Chr9 | 13433001 | A-G | HET | INTERGENIC | |
| SBS | Chr9 | 14978876 | G-T | HET | ||
| SBS | Chr9 | 2749621 | T-G | Homo | ||
| SBS | Chr9 | 2749621 | T-G | HOMO | INTRON | |
| SBS | Chr9 | 3016876 | T-C | HET | ||
| SBS | Chr9 | 3016876 | T-C | HET | INTRON | |
| SBS | Chr9 | 3164651 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os09g06660 |
| SBS | Chr9 | 7075511 | T-C | HET |
Deletions: 26
Insertions: 8
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 13679991 | 13679991 | 1 | |
| Insertion | Chr1 | 4555088 | 4555088 | 1 | |
| Insertion | Chr10 | 16392350 | 16392353 | 4 | |
| Insertion | Chr2 | 35077154 | 35077156 | 3 | |
| Insertion | Chr3 | 23942107 | 23942107 | 1 | |
| Insertion | Chr5 | 8116387 | 8116387 | 1 | LOC_Os05g14400 |
| Insertion | Chr8 | 17916507 | 17916507 | 1 | |
| Insertion | Chr9 | 11531490 | 11531490 | 1 |
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr1 | 39419949 | 39420120 |
Translocations: 16
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr10 | 164478 | Chr5 | 14966498 | |
| Translocation | Chr10 | 2694518 | Chr6 | 22683226 | |
| Translocation | Chr10 | 7833402 | Chr9 | 2382386 | 2 |
| Translocation | Chr9 | 8289918 | Chr6 | 11646591 | |
| Translocation | Chr3 | 15692309 | Chr2 | 18844091 | |
| Translocation | Chr12 | 18179682 | Chr11 | 10218713 | |
| Translocation | Chr10 | 18190003 | Chr4 | 8855441 | LOC_Os10g34078 |
| Translocation | Chr11 | 18371992 | Chr10 | 879842 | LOC_Os11g31440 |
| Translocation | Chr5 | 18580910 | Chr1 | 7861353 | |
| Translocation | Chr10 | 19291321 | Chr2 | 16123058 | |
| Translocation | Chr7 | 19416595 | Chr3 | 1603838 | |
| Translocation | Chr12 | 20175900 | Chr11 | 13411844 | LOC_Os12g33360 |
| Translocation | Chr11 | 22529369 | Chr8 | 10765832 | LOC_Os11g37990 |
| Translocation | Chr12 | 26966677 | Chr6 | 4303675 | |
| Translocation | Chr12 | 27044721 | Chr11 | 20669049 | |
| Translocation | Chr12 | 27530417 | Chr8 | 7729489 |
