Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2668-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2668-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 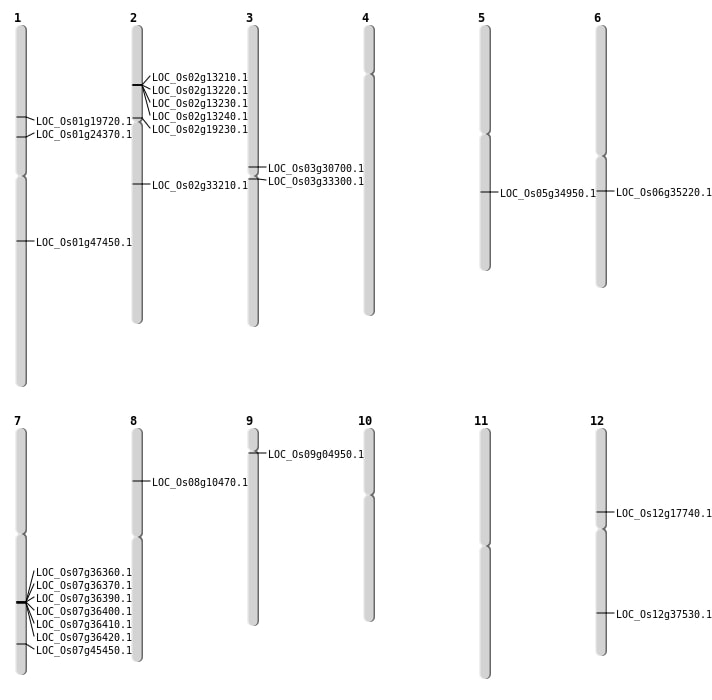
|
| Variant Information |
|---|
Single base substitutions: 53
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 11182700 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g19720 |
| SBS | Chr1 | 12013719 | A-G | HET | ||
| SBS | Chr1 | 12900153 | C-A | HET | ||
| SBS | Chr1 | 13784679 | C-T | HET | ||
| SBS | Chr1 | 15127295 | G-A | HET | ||
| SBS | Chr1 | 15127296 | C-A | HET | ||
| SBS | Chr1 | 22457931 | C-T | HET | ||
| SBS | Chr1 | 27119913 | G-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os01g47450 |
| SBS | Chr1 | 37383500 | G-T | HET | ||
| SBS | Chr1 | 40366243 | G-A | HET | ||
| SBS | Chr1 | 4533343 | G-T | HET | ||
| SBS | Chr10 | 1131478 | G-C | HET | ||
| SBS | Chr10 | 11677147 | G-A | HET | ||
| SBS | Chr10 | 12325811 | C-A | HET | ||
| SBS | Chr10 | 2501806 | T-C | HET | ||
| SBS | Chr10 | 7463387 | C-T | HET | ||
| SBS | Chr11 | 17858815 | G-A | HET | ||
| SBS | Chr12 | 10159353 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os12g17740 |
| SBS | Chr12 | 1483334 | C-T | HET | ||
| SBS | Chr12 | 22101250 | T-C | HET | ||
| SBS | Chr12 | 24484921 | T-A | HET | ||
| SBS | Chr2 | 13843421 | T-A | HET | ||
| SBS | Chr2 | 18742844 | C-T | Homo | ||
| SBS | Chr2 | 19743605 | G-A | HET | STOP_GAINED | LOC_Os02g33210 |
| SBS | Chr2 | 20405646 | T-A | HET | ||
| SBS | Chr2 | 20405647 | T-A | HET | ||
| SBS | Chr2 | 20406091 | T-G | HET | ||
| SBS | Chr2 | 22625218 | G-A | HET | ||
| SBS | Chr2 | 22685756 | A-T | HET | ||
| SBS | Chr2 | 24514837 | C-T | HET | ||
| SBS | Chr2 | 32100281 | C-T | HET | ||
| SBS | Chr2 | 7113972 | G-C | HET | ||
| SBS | Chr2 | 7113975 | G-A | HET | ||
| SBS | Chr2 | 7113979 | G-A | HET | ||
| SBS | Chr3 | 17490342 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g30700 |
| SBS | Chr3 | 18233471 | C-T | HET | ||
| SBS | Chr3 | 36054416 | G-A | Homo | ||
| SBS | Chr3 | 8263099 | T-C | HET | ||
| SBS | Chr4 | 19348645 | C-T | HET | ||
| SBS | Chr4 | 35233363 | G-A | HET | ||
| SBS | Chr5 | 26181041 | C-T | HET | ||
| SBS | Chr6 | 17726903 | C-G | HET | ||
| SBS | Chr6 | 9264118 | T-G | HET | ||
| SBS | Chr7 | 237306 | T-C | HET | ||
| SBS | Chr7 | 27050739 | A-G | HET | ||
| SBS | Chr7 | 27111278 | G-T | HET | STOP_GAINED | LOC_Os07g45450 |
| SBS | Chr8 | 15481145 | T-C | HET | ||
| SBS | Chr8 | 21369106 | G-A | HET | ||
| SBS | Chr8 | 26322774 | C-T | Homo | ||
| SBS | Chr9 | 12971796 | G-A | Homo | ||
| SBS | Chr9 | 19220275 | G-T | HET | ||
| SBS | Chr9 | 2657136 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os09g04950 |
| SBS | Chr9 | 7130343 | A-G | HET |
Deletions: 27
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr12 | 855933 | 855935 | 2 | |
| Deletion | Chr9 | 974019 | 974022 | 3 | |
| Deletion | Chr9 | 3360730 | 3360731 | 1 | |
| Deletion | Chr10 | 3643919 | 3643920 | 1 | |
| Deletion | Chr2 | 6956259 | 6956267 | 8 | |
| Deletion | Chr2 | 7035001 | 7056000 | 20999 | 4 |
| Deletion | Chr10 | 9573765 | 9573775 | 10 | |
| Deletion | Chr3 | 9802219 | 9802221 | 2 | |
| Deletion | Chr11 | 10971718 | 10971719 | 1 | |
| Deletion | Chr2 | 11224512 | 11224513 | 1 | LOC_Os02g19230 |
| Deletion | Chr6 | 14670930 | 14670937 | 7 | |
| Deletion | Chr2 | 17544798 | 17544799 | 1 | |
| Deletion | Chr12 | 19301303 | 19301315 | 12 | |
| Deletion | Chr6 | 20549335 | 20549336 | 1 | LOC_Os06g35220 |
| Deletion | Chr5 | 20762635 | 20762649 | 14 | LOC_Os05g34950 |
| Deletion | Chr5 | 21245043 | 21245044 | 1 | |
| Deletion | Chr7 | 21742001 | 21797000 | 54999 | 6 |
| Deletion | Chr12 | 22082587 | 22082597 | 10 | |
| Deletion | Chr3 | 22334453 | 22334484 | 31 | |
| Deletion | Chr12 | 23031797 | 23031803 | 6 | LOC_Os12g37530 |
| Deletion | Chr3 | 26316326 | 26316337 | 11 | |
| Deletion | Chr6 | 26955323 | 26955324 | 1 | |
| Deletion | Chr1 | 27467256 | 27467258 | 2 | |
| Deletion | Chr1 | 28106725 | 28106732 | 7 | |
| Deletion | Chr3 | 30380760 | 30380761 | 1 | |
| Deletion | Chr2 | 31132030 | 31132053 | 23 | |
| Deletion | Chr2 | 34710217 | 34710219 | 2 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr11 | 11558331 | 11558332 | 2 | |
| Insertion | Chr12 | 11417955 | 11417955 | 1 | |
| Insertion | Chr3 | 22607473 | 22607487 | 15 |
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr3 | 35758835 | 35759053 |
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr4 | 2929577 | Chr3 | 20535942 | |
| Translocation | Chr8 | 6156580 | Chr3 | 19048457 | 2 |
| Translocation | Chr8 | 6157001 | Chr3 | 19048457 | 2 |
| Translocation | Chr9 | 8398262 | Chr2 | 25035292 | |
| Translocation | Chr8 | 14865817 | Chr1 | 13740753 | 2 |
