Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2669-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2669-S Alignment File |
| Seed Availability | No |
| Mapping (Hover to Zoom-In) | 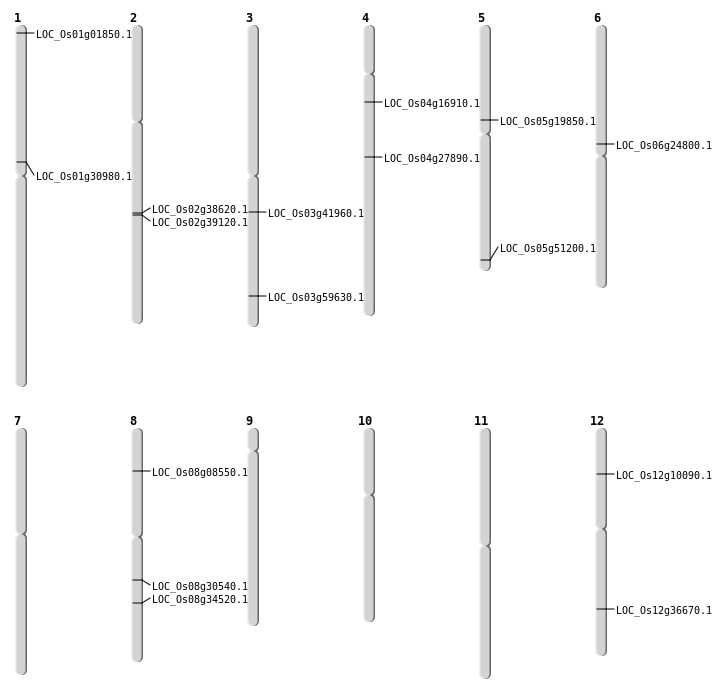
|
| Variant Information |
|---|
Single base substitutions: 41
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 10905318 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 16933074 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g30980 |
| SBS | Chr1 | 23299986 | G-C | HET | INTERGENIC | |
| SBS | Chr1 | 26741041 | A-T | HET | INTERGENIC | |
| SBS | Chr1 | 467591 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g01850 |
| SBS | Chr10 | 11521580 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 2580350 | A-G | HET | INTERGENIC | |
| SBS | Chr11 | 27266307 | A-G | HET | INTERGENIC | |
| SBS | Chr11 | 9642050 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 12483435 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 17334798 | G-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 10449090 | T-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 10891842 | C-A | HET | INTERGENIC | |
| SBS | Chr2 | 17739096 | A-T | HET | INTERGENIC | |
| SBS | Chr2 | 24883218 | G-C | HET | INTERGENIC | |
| SBS | Chr2 | 5757415 | G-A | HET | INTRON | |
| SBS | Chr3 | 18439267 | T-C | HOMO | INTERGENIC | |
| SBS | Chr3 | 18699014 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 201801 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 23315249 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g41960 |
| SBS | Chr3 | 33952328 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os03g59630 |
| SBS | Chr3 | 36191175 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 3000850 | C-A | HET | INTRON | |
| SBS | Chr4 | 3316310 | C-T | HET | INTRON | |
| SBS | Chr4 | 9249634 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os04g16910 |
| SBS | Chr5 | 11559291 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g19850 |
| SBS | Chr5 | 14870455 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 29370263 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g51200 |
| SBS | Chr5 | 5319256 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 14560063 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os06g24800 |
| SBS | Chr6 | 15303943 | C-A | HOMO | INTRON | |
| SBS | Chr6 | 19958316 | T-C | HOMO | INTRON | |
| SBS | Chr6 | 5276038 | T-C | HET | INTERGENIC | |
| SBS | Chr7 | 10938688 | C-A | HET | INTERGENIC | |
| SBS | Chr7 | 11557878 | A-T | HET | INTRON | |
| SBS | Chr7 | 12935761 | C-T | HET | INTRON | |
| SBS | Chr7 | 23696021 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 27582914 | C-T | HET | INTRON | |
| SBS | Chr8 | 10507488 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 18410445 | C-G | HET | INTERGENIC | |
| SBS | Chr8 | 4633708 | A-G | HET | INTRON |
Deletions: 30
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr6 | 1150471 | 1150480 | 9 | |
| Deletion | Chr8 | 1381500 | 1381501 | 1 | |
| Deletion | Chr4 | 1731385 | 1731387 | 2 | |
| Deletion | Chr2 | 2418968 | 2418969 | 1 | |
| Deletion | Chr1 | 3423610 | 3423615 | 5 | |
| Deletion | Chr5 | 3998081 | 3998090 | 9 | |
| Deletion | Chr9 | 4336918 | 4336919 | 1 | |
| Deletion | Chr12 | 9615535 | 9615628 | 93 | |
| Deletion | Chr11 | 11380109 | 11380110 | 1 | |
| Deletion | Chr8 | 11416088 | 11416093 | 5 | |
| Deletion | Chr12 | 11616289 | 11616290 | 1 | |
| Deletion | Chr1 | 12679750 | 12679752 | 2 | |
| Deletion | Chr7 | 12960193 | 12960194 | 1 | |
| Deletion | Chr9 | 13580714 | 13580715 | 1 | |
| Deletion | Chr9 | 16351084 | 16351093 | 9 | |
| Deletion | Chr2 | 17256557 | 17256565 | 8 | |
| Deletion | Chr7 | 18465279 | 18465280 | 1 | |
| Deletion | Chr8 | 18772001 | 18799000 | 26999 | 2 |
| Deletion | Chr2 | 19577666 | 19577669 | 3 | |
| Deletion | Chr8 | 21064793 | 21064794 | 1 | |
| Deletion | Chr2 | 21647614 | 21647618 | 4 | |
| Deletion | Chr1 | 22415418 | 22415422 | 4 | |
| Deletion | Chr12 | 22460491 | 22460504 | 13 | LOC_Os12g36670 |
| Deletion | Chr2 | 22702105 | 22702107 | 2 | |
| Deletion | Chr3 | 23254062 | 23254073 | 11 | |
| Deletion | Chr2 | 23627297 | 23627298 | 1 | LOC_Os02g39120 |
| Deletion | Chr8 | 24349849 | 24349850 | 1 | |
| Deletion | Chr12 | 24606412 | 24606430 | 18 | |
| Deletion | Chr4 | 27054128 | 27054129 | 1 | |
| Deletion | Chr1 | 28241028 | 28241034 | 6 |
Insertions: 10
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 40519340 | 40519383 | 44 | |
| Insertion | Chr12 | 5343305 | 5343306 | 2 | LOC_Os12g10090 |
| Insertion | Chr2 | 21223671 | 21223671 | 1 | |
| Insertion | Chr3 | 23901198 | 23901198 | 1 | |
| Insertion | Chr4 | 16473545 | 16473549 | 5 | LOC_Os04g27890 |
| Insertion | Chr5 | 8256416 | 8256416 | 1 | |
| Insertion | Chr6 | 9639759 | 9639802 | 44 | |
| Insertion | Chr8 | 18788919 | 18788919 | 1 | |
| Insertion | Chr9 | 3093750 | 3093761 | 12 | |
| Insertion | Chr9 | 5950876 | 5950879 | 4 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr10 | 16478869 | 17124054 | |
| Inversion | Chr10 | 16479154 | 17124055 |
Translocations: 6
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr8 | 2675333 | Chr2 | 3777780 | |
| Translocation | Chr8 | 4935346 | Chr2 | 23342788 | 2 |
| Translocation | Chr11 | 10341713 | Chr10 | 16478869 | |
| Translocation | Chr11 | 10341715 | Chr10 | 16479149 | |
| Translocation | Chr6 | 19268925 | Chr1 | 15459829 | |
| Translocation | Chr4 | 34580583 | Chr1 | 2756226 |
