Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2673-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2673-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 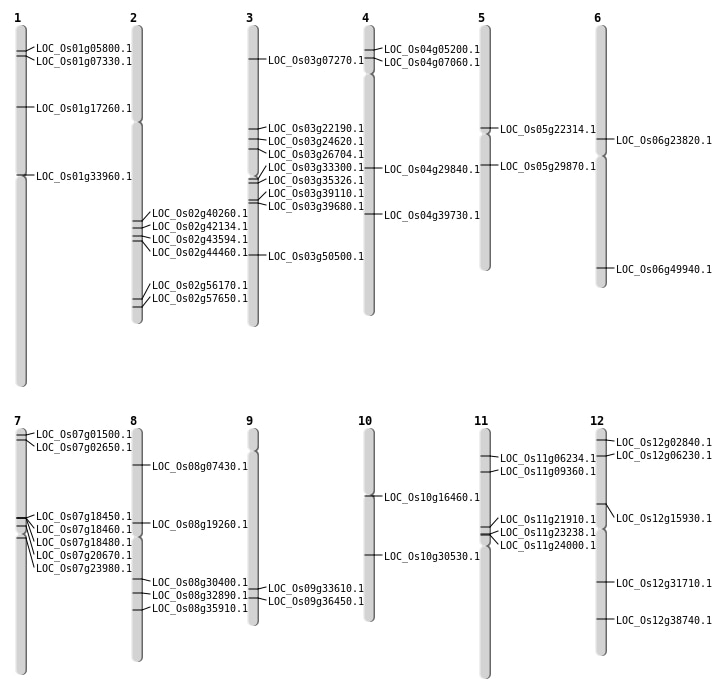
|
| Variant Information |
|---|
Single base substitutions: 83
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 20013569 | A-G | HET | ||
| SBS | Chr1 | 21867715 | G-A | Homo | ||
| SBS | Chr1 | 22375233 | G-T | HET | ||
| SBS | Chr1 | 22924920 | G-A | Homo | ||
| SBS | Chr1 | 33081710 | A-G | HET | ||
| SBS | Chr1 | 38836741 | T-A | HET | ||
| SBS | Chr1 | 9644959 | C-G | HET | ||
| SBS | Chr10 | 9510491 | T-C | Homo | ||
| SBS | Chr11 | 1392160 | G-T | HET | ||
| SBS | Chr11 | 15112535 | G-T | HET | ||
| SBS | Chr11 | 22950737 | G-A | HET | ||
| SBS | Chr11 | 25350641 | A-G | HET | ||
| SBS | Chr11 | 28681757 | G-A | HET | ||
| SBS | Chr11 | 4563823 | C-T | HET | ||
| SBS | Chr11 | 4730273 | G-A | HET | ||
| SBS | Chr11 | 8352051 | T-C | HET | ||
| SBS | Chr12 | 1019773 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os12g02840 |
| SBS | Chr12 | 17227670 | C-A | HET | ||
| SBS | Chr12 | 8150775 | G-A | HET | ||
| SBS | Chr12 | 9083682 | C-G | Homo | NON_SYNONYMOUS_CODING | LOC_Os12g15930 |
| SBS | Chr2 | 115824 | A-T | HET | ||
| SBS | Chr2 | 1204777 | G-A | HET | ||
| SBS | Chr2 | 192402 | G-T | HET | ||
| SBS | Chr2 | 21085746 | G-A | HET | ||
| SBS | Chr2 | 33207398 | T-A | Homo | ||
| SBS | Chr2 | 34363712 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g56170 |
| SBS | Chr2 | 3504081 | G-A | HET | ||
| SBS | Chr2 | 7579652 | T-A | HET | ||
| SBS | Chr2 | 7980098 | G-A | Homo | ||
| SBS | Chr3 | 11677125 | C-T | HET | ||
| SBS | Chr3 | 12725963 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g22190 |
| SBS | Chr3 | 14022876 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os03g24620 |
| SBS | Chr3 | 17122078 | C-T | HET | ||
| SBS | Chr3 | 19046902 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os03g33300 |
| SBS | Chr3 | 19085781 | T-C | HET | ||
| SBS | Chr3 | 22486977 | G-C | HET | ||
| SBS | Chr3 | 23303767 | T-C | HET | ||
| SBS | Chr3 | 26064342 | A-T | HET | ||
| SBS | Chr3 | 27700727 | A-T | HET | ||
| SBS | Chr3 | 28831727 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g50500 |
| SBS | Chr3 | 29269178 | T-A | HET | ||
| SBS | Chr3 | 29269204 | C-T | HET | ||
| SBS | Chr3 | 33274291 | A-G | HET | ||
| SBS | Chr3 | 6211312 | T-C | Homo | ||
| SBS | Chr4 | 13082613 | G-A | HET | ||
| SBS | Chr4 | 21589457 | G-A | HET | ||
| SBS | Chr4 | 23677604 | C-A | HET | STOP_GAINED | LOC_Os04g39730 |
| SBS | Chr4 | 23695555 | G-A | HET | ||
| SBS | Chr4 | 26239678 | A-T | Homo | ||
| SBS | Chr4 | 28545761 | A-T | HET | ||
| SBS | Chr4 | 33216957 | G-A | HET | ||
| SBS | Chr4 | 6624937 | C-T | HET | ||
| SBS | Chr5 | 11506852 | G-T | HET | ||
| SBS | Chr5 | 17284382 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os05g29870 |
| SBS | Chr5 | 18221682 | T-C | Homo | ||
| SBS | Chr5 | 19061077 | A-G | HET | ||
| SBS | Chr5 | 19130123 | C-T | HET | ||
| SBS | Chr5 | 20202865 | A-G | HET | ||
| SBS | Chr5 | 22654220 | C-T | HET | ||
| SBS | Chr5 | 3434030 | T-C | HET | ||
| SBS | Chr5 | 4549729 | G-A | HET | ||
| SBS | Chr5 | 5790095 | G-T | HET | ||
| SBS | Chr5 | 7956566 | A-G | HET | ||
| SBS | Chr6 | 10831119 | G-A | HET | ||
| SBS | Chr6 | 13924830 | T-C | Homo | NON_SYNONYMOUS_CODING | LOC_Os06g23820 |
| SBS | Chr6 | 30235925 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os06g49940 |
| SBS | Chr7 | 10938553 | T-A | Homo | ||
| SBS | Chr7 | 12300123 | C-T | HET | ||
| SBS | Chr7 | 14292636 | G-T | HET | ||
| SBS | Chr7 | 1458235 | C-T | Homo | ||
| SBS | Chr7 | 316921 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g01500 |
| SBS | Chr8 | 11749591 | C-T | Homo | ||
| SBS | Chr8 | 18701274 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os08g30400 |
| SBS | Chr8 | 20397858 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g32890 |
| SBS | Chr8 | 25376111 | A-G | Homo | ||
| SBS | Chr8 | 4049873 | G-A | HET | ||
| SBS | Chr8 | 4211352 | T-A | HET | ||
| SBS | Chr9 | 1069589 | C-G | HET | ||
| SBS | Chr9 | 11242411 | G-A | HET | ||
| SBS | Chr9 | 21034729 | T-C | HET | ||
| SBS | Chr9 | 2121420 | A-G | HET | ||
| SBS | Chr9 | 22784991 | G-T | HET | ||
| SBS | Chr9 | 8378227 | T-C | HET |
Deletions: 20
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr4 | 2594877 | 2594881 | 4 | LOC_Os04g05200 |
| Deletion | Chr11 | 2983758 | 2983764 | 6 | LOC_Os11g06234 |
| Deletion | Chr8 | 5515735 | 5515747 | 12 | |
| Deletion | Chr10 | 7345299 | 7345307 | 8 | |
| Deletion | Chr12 | 9762954 | 9762956 | 2 | |
| Deletion | Chr7 | 10911001 | 10937000 | 25999 | 3 |
| Deletion | Chr5 | 11565505 | 11565507 | 2 | |
| Deletion | Chr11 | 12578428 | 12578435 | 7 | LOC_Os11g21910 |
| Deletion | Chr7 | 12629799 | 12629828 | 29 | |
| Deletion | Chr2 | 15668934 | 15668973 | 39 | |
| Deletion | Chr10 | 15874500 | 15874503 | 3 | LOC_Os10g30530 |
| Deletion | Chr10 | 16109107 | 16109112 | 5 | |
| Deletion | Chr11 | 18939463 | 18939464 | 1 | |
| Deletion | Chr11 | 19896546 | 19896567 | 21 | |
| Deletion | Chr9 | 21770867 | 21770868 | 1 | |
| Deletion | Chr1 | 23862903 | 23863398 | 495 | |
| Deletion | Chr8 | 25407453 | 25407454 | 1 | |
| Deletion | Chr6 | 25969971 | 25969983 | 12 | |
| Deletion | Chr3 | 28176711 | 28176713 | 2 | |
| Deletion | Chr11 | 28591618 | 28591619 | 1 |
Insertions: 7
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 4874203 | 4874203 | 1 | |
| Insertion | Chr3 | 32641582 | 32641583 | 2 | |
| Insertion | Chr4 | 3694899 | 3694902 | 4 | LOC_Os04g07060 |
| Insertion | Chr4 | 5677293 | 5677296 | 4 | |
| Insertion | Chr6 | 15437370 | 15437370 | 1 | |
| Insertion | Chr6 | 8810277 | 8810277 | 1 | |
| Insertion | Chr7 | 7882486 | 7882486 | 1 |
Inversions: 7
Translocations: 43
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr2 | 2377045 | Chr1 | 2332548 | |
| Translocation | Chr12 | 2966113 | Chr2 | 3277505 | LOC_Os12g06230 |
| Translocation | Chr11 | 3435412 | Chr1 | 30237026 | |
| Translocation | Chr8 | 4172940 | Chr6 | 7544380 | LOC_Os08g07430 |
| Translocation | Chr6 | 4273614 | Chr3 | 27840704 | |
| Translocation | Chr11 | 5027349 | Chr1 | 3456694 | 2 |
| Translocation | Chr9 | 6282553 | Chr4 | 5870526 | |
| Translocation | Chr12 | 6457494 | Chr4 | 2769002 | |
| Translocation | Chr11 | 6551189 | Chr3 | 21439963 | |
| Translocation | Chr11 | 6668152 | Chr7 | 958499 | 2 |
| Translocation | Chr10 | 8207628 | Chr7 | 11938715 | 2 |
| Translocation | Chr9 | 9104328 | Chr4 | 17791425 | 2 |
| Translocation | Chr9 | 9116870 | Chr8 | 16818357 | |
| Translocation | Chr6 | 9219394 | Chr3 | 14907255 | |
| Translocation | Chr12 | 9955132 | Chr1 | 39125737 | |
| Translocation | Chr9 | 10934374 | Chr1 | 8783442 | |
| Translocation | Chr8 | 11520560 | Chr2 | 35304210 | 2 |
| Translocation | Chr7 | 11938655 | Chr1 | 16975542 | LOC_Os07g20670 |
| Translocation | Chr7 | 11938723 | Chr2 | 13600035 | LOC_Os07g20670 |
| Translocation | Chr4 | 12370201 | Chr2 | 25329650 | 2 |
| Translocation | Chr7 | 13573678 | Chr5 | 12627012 | 2 |
| Translocation | Chr10 | 15469240 | Chr2 | 26917497 | 2 |
| Translocation | Chr8 | 15765976 | Chr3 | 15248051 | 2 |
| Translocation | Chr2 | 16599749 | Chr1 | 19738564 | |
| Translocation | Chr12 | 16959656 | Chr2 | 611354 | |
| Translocation | Chr7 | 17052497 | Chr1 | 2773981 | 2 |
| Translocation | Chr12 | 19090671 | Chr10 | 1076403 | LOC_Os12g31710 |
| Translocation | Chr3 | 19544628 | Chr1 | 9929654 | 2 |
| Translocation | Chr9 | 19846048 | Chr1 | 3204859 | LOC_Os09g33610 |
| Translocation | Chr12 | 20000154 | Chr2 | 20513525 | |
| Translocation | Chr9 | 20294565 | Chr1 | 33161280 | |
| Translocation | Chr7 | 21033253 | Chr5 | 26268341 | |
| Translocation | Chr7 | 22081676 | Chr4 | 16289132 | |
| Translocation | Chr8 | 22651596 | Chr7 | 3871170 | LOC_Os08g35910 |
| Translocation | Chr8 | 22803825 | Chr7 | 1478338 | |
| Translocation | Chr6 | 23042478 | Chr3 | 3707349 | 2 |
| Translocation | Chr12 | 23803489 | Chr3 | 22076017 | 2 |
| Translocation | Chr12 | 24279596 | Chr2 | 26313990 | 2 |
| Translocation | Chr4 | 25756767 | Chr1 | 2816738 | |
| Translocation | Chr4 | 27248012 | Chr2 | 24373941 | 2 |
| Translocation | Chr7 | 28085274 | Chr1 | 15882272 | |
| Translocation | Chr3 | 28718267 | Chr1 | 18688725 | 2 |
| Translocation | Chr4 | 32281557 | Chr3 | 21730919 | 2 |
