Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2687-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2687-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 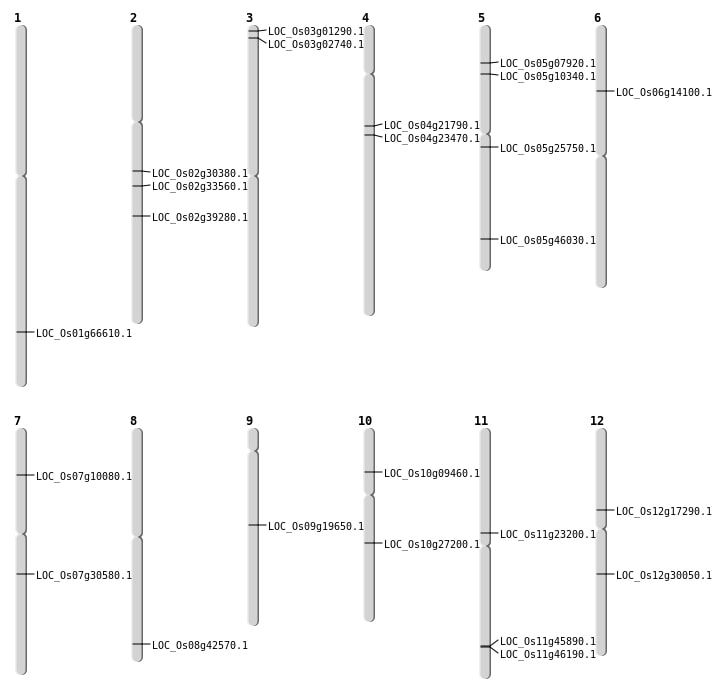
|
| Variant Information |
|---|
Single base substitutions: 82
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 18713288 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 33628472 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 34890123 | A-T | HET | INTRON | |
| SBS | Chr1 | 6098068 | C-T | Homo | ||
| SBS | Chr1 | 6969175 | C-T | HET | INTRON | |
| SBS | Chr1 | 741849 | A-G | HET | ||
| SBS | Chr10 | 8411033 | G-T | HET | ||
| SBS | Chr10 | 9539218 | C-A | HET | ||
| SBS | Chr11 | 16493950 | C-A | HET | ||
| SBS | Chr11 | 16493950 | C-A | HET | INTERGENIC | |
| SBS | Chr11 | 16493951 | G-A | HET | ||
| SBS | Chr11 | 16493951 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 25247462 | A-G | HET | ||
| SBS | Chr11 | 29001206 | G-T | HET | ||
| SBS | Chr11 | 29001206 | G-T | HET | INTERGENIC | |
| SBS | Chr11 | 6477110 | A-T | Homo | ||
| SBS | Chr11 | 6477110 | A-T | HOMO | INTRON | |
| SBS | Chr11 | 7414075 | T-C | HET | ||
| SBS | Chr12 | 13031360 | A-T | HET | ||
| SBS | Chr12 | 13031360 | A-T | HET | INTERGENIC | |
| SBS | Chr12 | 23541166 | C-A | HET | ||
| SBS | Chr12 | 24449715 | G-A | Homo | ||
| SBS | Chr12 | 24449715 | G-A | HOMO | INTERGENIC | |
| SBS | Chr12 | 9893018 | G-A | HET | STOP_GAINED | LOC_Os12g17290 |
| SBS | Chr2 | 18092046 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os02g30380 |
| SBS | Chr2 | 30212386 | T-C | HET | ||
| SBS | Chr2 | 30212386 | T-C | HET | INTERGENIC | |
| SBS | Chr2 | 30782225 | C-T | HET | ||
| SBS | Chr3 | 12170095 | C-T | Homo | ||
| SBS | Chr3 | 12170095 | C-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 6473224 | T-C | HET | ||
| SBS | Chr3 | 6473224 | T-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr4 | 12343611 | A-C | HET | NON_SYNONYMOUS_CODING | LOC_Os04g21790 |
| SBS | Chr4 | 13834097 | T-C | HET | ||
| SBS | Chr4 | 16551540 | C-T | HET | ||
| SBS | Chr4 | 16551540 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 20033703 | A-G | HET | ||
| SBS | Chr4 | 20033703 | A-G | HET | INTERGENIC | |
| SBS | Chr4 | 20033712 | G-A | HET | ||
| SBS | Chr4 | 20033712 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 27923067 | G-T | HET | ||
| SBS | Chr4 | 27923067 | G-T | HET | INTRON | |
| SBS | Chr4 | 5790280 | C-T | HET | ||
| SBS | Chr5 | 14297594 | G-T | HET | ||
| SBS | Chr5 | 14980319 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os05g25750 |
| SBS | Chr5 | 20768500 | G-A | HET | ||
| SBS | Chr5 | 21101966 | G-T | HET | INTERGENIC | |
| SBS | Chr5 | 27889063 | T-G | Homo | ||
| SBS | Chr5 | 27889063 | T-G | HOMO | SPLICE_SITE_REGION | |
| SBS | Chr5 | 3735695 | T-A | HET | ||
| SBS | Chr5 | 3735695 | T-A | HET | INTERGENIC | |
| SBS | Chr6 | 4154261 | A-T | HET | ||
| SBS | Chr6 | 4154261 | A-T | HET | INTERGENIC | |
| SBS | Chr6 | 4876222 | G-T | HET | INTRON | |
| SBS | Chr6 | 5888138 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 11293103 | C-T | HET | ||
| SBS | Chr7 | 18095552 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g30580 |
| SBS | Chr7 | 18095553 | A-C | HET | NON_SYNONYMOUS_CODING | LOC_Os07g30580 |
| SBS | Chr7 | 18963511 | G-C | HET | ||
| SBS | Chr7 | 18963511 | G-C | HET | INTERGENIC | |
| SBS | Chr7 | 19153135 | C-T | HET | ||
| SBS | Chr7 | 24198867 | C-T | HET | ||
| SBS | Chr7 | 25003273 | A-T | HET | ||
| SBS | Chr7 | 27925150 | C-A | HET | ||
| SBS | Chr7 | 27925150 | C-A | HET | INTERGENIC | |
| SBS | Chr7 | 6069413 | A-G | HET | ||
| SBS | Chr7 | 6069413 | A-G | HET | INTERGENIC | |
| SBS | Chr8 | 13976246 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 1399006 | A-T | HET | ||
| SBS | Chr8 | 26169272 | G-C | Homo | ||
| SBS | Chr8 | 26169272 | G-C | HOMO | INTERGENIC | |
| SBS | Chr8 | 7272318 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 11748998 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os09g19650 |
| SBS | Chr9 | 13833451 | G-A | Homo | ||
| SBS | Chr9 | 13833451 | G-A | HOMO | UTR_3_PRIME | |
| SBS | Chr9 | 16824334 | G-T | Homo | ||
| SBS | Chr9 | 16824334 | G-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr9 | 17058830 | G-A | HET | ||
| SBS | Chr9 | 19130043 | G-T | HET | ||
| SBS | Chr9 | 7333416 | T-G | HET | ||
| SBS | Chr9 | 7333416 | T-G | HET | INTERGENIC | |
| SBS | Chr9 | 8276632 | C-T | HET |
Deletions: 31
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr3 | 614294 | 614296 | 2 | |
| Deletion | Chr3 | 1028215 | 1028227 | 12 | 2 |
| Deletion | Chr2 | 2403696 | 2403703 | 7 | |
| Deletion | Chr3 | 2648685 | 2648706 | 21 | |
| Deletion | Chr3 | 3707272 | 3707273 | 1 | |
| Deletion | Chr10 | 5101993 | 5101999 | 6 | LOC_Os10g09460 |
| Deletion | Chr7 | 5395845 | 5395861 | 16 | LOC_Os07g10080 |
| Deletion | Chr12 | 5633839 | 5633865 | 26 | |
| Deletion | Chr9 | 10443317 | 10443318 | 1 | |
| Deletion | Chr7 | 11816198 | 11816199 | 1 | |
| Deletion | Chr2 | 12259160 | 12259161 | 1 | |
| Deletion | Chr9 | 12680848 | 12680849 | 1 | 2 |
| Deletion | Chr12 | 12916543 | 12916584 | 41 | |
| Deletion | Chr9 | 13461928 | 13461929 | 1 | 2 |
| Deletion | Chr4 | 15182299 | 15182300 | 1 | |
| Deletion | Chr11 | 15722872 | 15722873 | 1 | |
| Deletion | Chr10 | 15923379 | 15923385 | 6 | 2 |
| Deletion | Chr6 | 17764103 | 17764104 | 1 | |
| Deletion | Chr12 | 17996187 | 17996188 | 1 | LOC_Os12g30050 |
| Deletion | Chr3 | 21391561 | 21391568 | 7 | 2 |
| Deletion | Chr5 | 22941557 | 22941561 | 4 | 2 |
| Deletion | Chr12 | 23748961 | 23748962 | 1 | |
| Deletion | Chr5 | 26587739 | 26587740 | 1 | 2 |
| Deletion | Chr5 | 26693302 | 26693303 | 1 | 2 |
| Deletion | Chr11 | 27974802 | 27974826 | 24 | LOC_Os11g46190 |
| Deletion | Chr7 | 28176929 | 28176930 | 1 | |
| Deletion | Chr7 | 29086688 | 29086689 | 1 | |
| Deletion | Chr2 | 30173658 | 30173659 | 1 | |
| Deletion | Chr3 | 33842319 | 33842326 | 7 | |
| Deletion | Chr1 | 38688619 | 38688628 | 9 | LOC_Os01g66610 |
| Deletion | Chr1 | 39713093 | 39713095 | 2 |
Insertions: 9
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr10 | 9807979 | 9807979 | 1 | |
| Insertion | Chr11 | 13356703 | 13356703 | 1 | LOC_Os11g23200 |
| Insertion | Chr2 | 14810340 | 14810340 | 1 | |
| Insertion | Chr2 | 23808168 | 23808168 | 1 | |
| Insertion | Chr3 | 15780243 | 15780244 | 2 | |
| Insertion | Chr5 | 2289603 | 2289610 | 8 | |
| Insertion | Chr7 | 14901434 | 14901434 | 1 | |
| Insertion | Chr8 | 26716485 | 26716486 | 2 | 2 |
| Insertion | Chr8 | 26716485 | 26716486 | 2 | 2 |
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr3 | 204355 | 588327 | 2 |
| Inversion | Chr3 | 204357 | 588329 | 2 |
| Inversion | Chr1 | 3966516 | 3966644 | |
| Inversion | Chr11 | 11867736 | 11867927 |
Translocations: 23
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 898529 | Chr4 | 17915299 | |
| Translocation | Chr9 | 1269806 | Chr3 | 18058077 | |
| Translocation | Chr10 | 3120137 | Chr1 | 23590110 | |
| Translocation | Chr5 | 4292257 | Chr4 | 13415270 | 2 |
| Translocation | Chr12 | 6248354 | Chr8 | 4190745 | |
| Translocation | Chr10 | 14333309 | Chr9 | 10527948 | LOC_Os10g27200 |
| Translocation | Chr11 | 15401580 | Chr4 | 18466158 | |
| Translocation | Chr10 | 16401320 | Chr2 | 19964529 | 2 |
| Translocation | Chr6 | 16817203 | Chr1 | 29886717 | |
| Translocation | Chr6 | 18884707 | Chr5 | 5645992 | LOC_Os05g10340 |
| Translocation | Chr7 | 19416460 | Chr1 | 34821287 | |
| Translocation | Chr4 | 20478519 | Chr3 | 26496416 | |
| Translocation | Chr12 | 20535474 | Chr1 | 34375468 | |
| Translocation | Chr12 | 21737271 | Chr6 | 7862478 | 2 |
| Translocation | Chr6 | 22827900 | Chr5 | 15579833 | |
| Translocation | Chr2 | 23723930 | Chr1 | 14275920 | LOC_Os02g39280 |
| Translocation | Chr12 | 24393553 | Chr6 | 19207692 | |
| Translocation | Chr8 | 24683903 | Chr1 | 39127474 | |
| Translocation | Chr11 | 24994225 | Chr4 | 17874176 | |
| Translocation | Chr7 | 25285592 | Chr2 | 13402391 | |
| Translocation | Chr8 | 26906514 | Chr3 | 25348842 | LOC_Os08g42570 |
| Translocation | Chr11 | 27570450 | Chr5 | 16490148 | |
| Translocation | Chr11 | 27761200 | Chr7 | 26193088 | LOC_Os11g45890 |
