Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2696-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2696-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 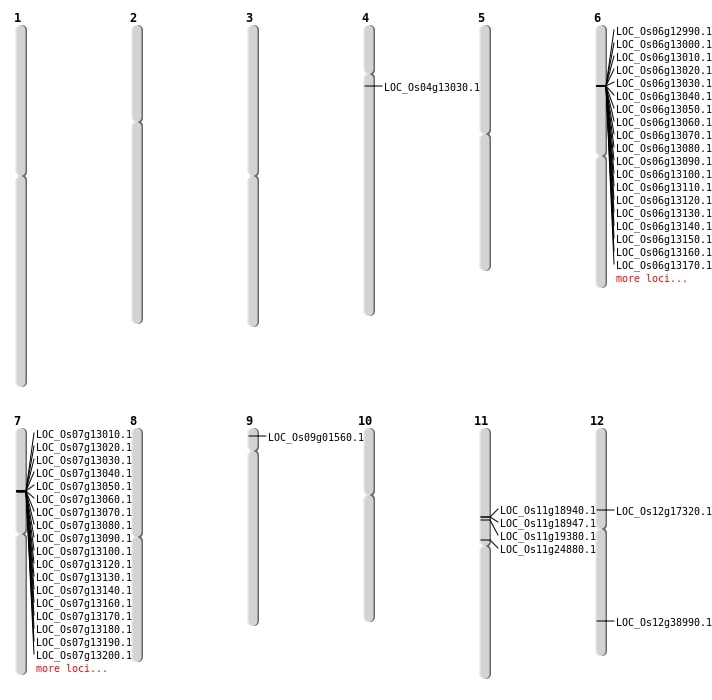
|
| Variant Information |
|---|
Single base substitutions: 61
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 13907033 | C-A | HET | ||
| SBS | Chr1 | 19177014 | T-C | HET | ||
| SBS | Chr1 | 20113792 | C-T | Homo | ||
| SBS | Chr1 | 24395283 | G-T | HET | ||
| SBS | Chr1 | 25028924 | A-T | HET | ||
| SBS | Chr1 | 32729826 | C-A | HET | ||
| SBS | Chr1 | 42111458 | G-T | Homo | ||
| SBS | Chr10 | 12101135 | G-A | HET | ||
| SBS | Chr10 | 13147824 | A-G | HET | ||
| SBS | Chr10 | 20842364 | C-A | HET | ||
| SBS | Chr10 | 21433719 | G-A | HET | ||
| SBS | Chr10 | 5184816 | C-T | HET | ||
| SBS | Chr10 | 6532447 | G-C | HET | ||
| SBS | Chr10 | 7352155 | C-T | HET | ||
| SBS | Chr11 | 10923621 | C-G | HET | ||
| SBS | Chr11 | 11151639 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os11g19380 |
| SBS | Chr11 | 18635179 | T-C | HET | ||
| SBS | Chr11 | 20614644 | T-C | HET | ||
| SBS | Chr11 | 22839130 | C-T | Homo | ||
| SBS | Chr11 | 23523805 | T-A | HET | ||
| SBS | Chr11 | 9043345 | T-A | HET | ||
| SBS | Chr12 | 1057562 | T-C | Homo | ||
| SBS | Chr12 | 14104488 | G-A | HET | ||
| SBS | Chr12 | 1880528 | C-A | HET | ||
| SBS | Chr12 | 19323116 | C-A | HET | ||
| SBS | Chr12 | 23980895 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os12g38990 |
| SBS | Chr2 | 26441635 | C-T | Homo | ||
| SBS | Chr2 | 31325573 | C-T | Homo | ||
| SBS | Chr3 | 19403080 | C-T | HET | ||
| SBS | Chr3 | 19673080 | G-A | HET | ||
| SBS | Chr3 | 32726847 | C-T | HET | ||
| SBS | Chr3 | 35894073 | C-A | HET | ||
| SBS | Chr4 | 18517858 | C-T | HET | ||
| SBS | Chr4 | 20980275 | T-G | HET | ||
| SBS | Chr4 | 21174321 | T-C | HET | ||
| SBS | Chr4 | 2292357 | C-T | HET | ||
| SBS | Chr4 | 24208331 | A-G | HET | ||
| SBS | Chr4 | 29283180 | C-T | HET | ||
| SBS | Chr4 | 33123656 | T-C | Homo | ||
| SBS | Chr4 | 7184219 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g13030 |
| SBS | Chr4 | 9402747 | T-A | HET | ||
| SBS | Chr5 | 13305800 | G-A | HET | ||
| SBS | Chr5 | 18649326 | C-T | HET | ||
| SBS | Chr5 | 27600410 | C-A | HET | ||
| SBS | Chr6 | 17232918 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os06g29920 |
| SBS | Chr6 | 30198051 | T-A | HET | ||
| SBS | Chr6 | 30624971 | T-C | HET | ||
| SBS | Chr6 | 5146983 | G-A | HET | ||
| SBS | Chr6 | 6250718 | C-T | HET | ||
| SBS | Chr6 | 6382052 | A-T | HET | ||
| SBS | Chr6 | 7315505 | C-T | HET | ||
| SBS | Chr7 | 17999662 | G-A | HET | ||
| SBS | Chr7 | 23019879 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os07g38320 |
| SBS | Chr7 | 27542539 | A-G | HET | ||
| SBS | Chr8 | 12053810 | A-G | Homo | ||
| SBS | Chr8 | 202429 | T-C | HET | ||
| SBS | Chr8 | 23800145 | T-A | HET | ||
| SBS | Chr8 | 8418751 | G-T | HET | ||
| SBS | Chr9 | 11044317 | T-A | HET | ||
| SBS | Chr9 | 14289727 | G-T | HET | ||
| SBS | Chr9 | 397612 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os09g01560 |
Deletions: 21
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr10 | 1097086 | 1097088 | 2 | |
| Deletion | Chr9 | 2332625 | 2332626 | 1 | |
| Deletion | Chr5 | 2512260 | 2512277 | 17 | |
| Deletion | Chr10 | 5712533 | 5712534 | 1 | |
| Deletion | Chr1 | 6977697 | 6977698 | 1 | |
| Deletion | Chr6 | 7112001 | 7230000 | 117999 | 20 |
| Deletion | Chr6 | 7423001 | 7471000 | 47999 | 7 |
| Deletion | Chr7 | 7456001 | 7739000 | 282999 | 45 |
| Deletion | Chr10 | 8253274 | 8253275 | 1 | |
| Deletion | Chr10 | 9901085 | 9901087 | 2 | |
| Deletion | Chr11 | 10767001 | 10796000 | 28999 | 2 |
| Deletion | Chr12 | 13438572 | 13438589 | 17 | |
| Deletion | Chr11 | 14180753 | 14180758 | 5 | LOC_Os11g24880 |
| Deletion | Chr5 | 14325195 | 14325231 | 36 | |
| Deletion | Chr10 | 15261004 | 15261007 | 3 | |
| Deletion | Chr11 | 16486271 | 16486274 | 3 | |
| Deletion | Chr4 | 16753460 | 16753461 | 1 | |
| Deletion | Chr10 | 20202369 | 20202395 | 26 | |
| Deletion | Chr4 | 23197288 | 23197290 | 2 | |
| Deletion | Chr2 | 34375954 | 34375955 | 1 | |
| Deletion | Chr1 | 38039606 | 38039620 | 14 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr2 | 32487529 | 32487530 | 2 | |
| Insertion | Chr3 | 12881950 | 12881952 | 3 | |
| Insertion | Chr8 | 7130239 | 7130239 | 1 |
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr6 | 9402720 | 9403593 | LOC_Os06g16430 |
| Inversion | Chr6 | 9402854 | 9403593 | LOC_Os06g16430 |
| Inversion | Chr12 | 9915303 | 9954710 | LOC_Os12g17320 |
| Inversion | Chr12 | 9915304 | 9954712 | LOC_Os12g17320 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr8 | 392592 | Chr6 | 9402844 | 2 |
| Translocation | Chr8 | 392593 | Chr6 | 9402735 | 2 |
| Translocation | Chr7 | 17141775 | Chr6 | 16283195 | LOC_Os07g29240 |
