Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2704-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2704-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 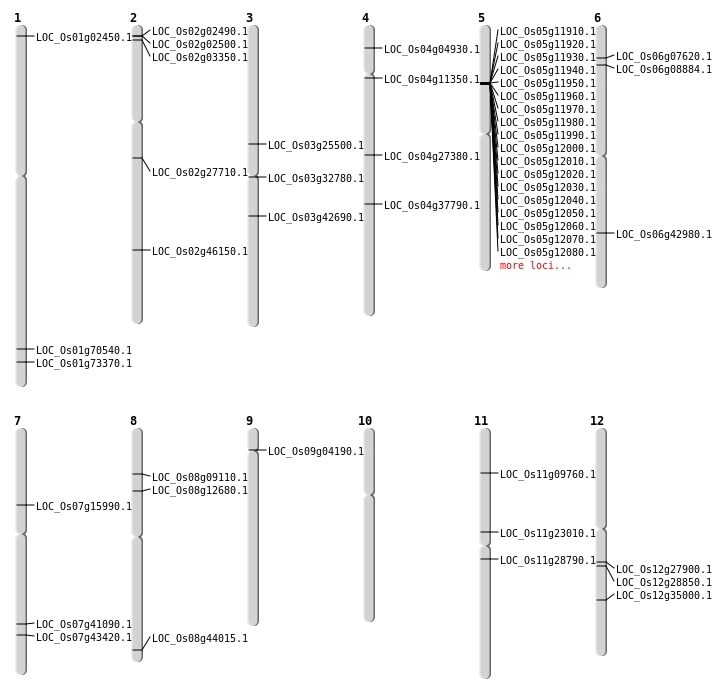
|
| Variant Information |
|---|
Single base substitutions: 100
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 23653042 | G-A | Homo | ||
| SBS | Chr1 | 23653042 | G-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 24884452 | A-T | HET | INTERGENIC | |
| SBS | Chr1 | 34490138 | C-A | HET | INTERGENIC | |
| SBS | Chr1 | 38487470 | C-A | HET | INTERGENIC | |
| SBS | Chr1 | 38799847 | C-T | HET | ||
| SBS | Chr1 | 38799847 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 40085038 | T-C | HET | ||
| SBS | Chr1 | 40852581 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g70540 |
| SBS | Chr1 | 793267 | T-A | HET | STOP_GAINED | LOC_Os01g02450 |
| SBS | Chr10 | 12689208 | T-A | Homo | ||
| SBS | Chr10 | 12689208 | T-A | HOMO | INTERGENIC | |
| SBS | Chr10 | 15468601 | C-G | HOMO | INTERGENIC | |
| SBS | Chr10 | 15795952 | C-T | HET | ||
| SBS | Chr11 | 11746296 | G-T | HET | INTRON | |
| SBS | Chr11 | 14332209 | C-T | HET | ||
| SBS | Chr11 | 6182169 | A-G | HET | ||
| SBS | Chr11 | 6182169 | A-G | HET | INTRON | |
| SBS | Chr12 | 10662318 | C-T | HET | ||
| SBS | Chr12 | 12617631 | C-T | HET | ||
| SBS | Chr12 | 17053613 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g28850 |
| SBS | Chr12 | 19791551 | T-A | HET | ||
| SBS | Chr12 | 19791551 | T-A | HET | INTERGENIC | |
| SBS | Chr12 | 20492351 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 21305855 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os12g35000 |
| SBS | Chr12 | 21509476 | C-A | HET | ||
| SBS | Chr12 | 7015107 | T-C | HET | INTERGENIC | |
| SBS | Chr2 | 12893095 | G-A | HET | ||
| SBS | Chr2 | 12893095 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 12893096 | C-T | HET | ||
| SBS | Chr2 | 12893096 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 16415375 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os02g27710 |
| SBS | Chr2 | 6550729 | T-C | Homo | ||
| SBS | Chr2 | 6550729 | T-C | HOMO | INTERGENIC | |
| SBS | Chr3 | 14567552 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os03g25500 |
| SBS | Chr3 | 18763386 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g32780 |
| SBS | Chr3 | 19532254 | A-G | HET | ||
| SBS | Chr3 | 19532254 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 23776295 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g42690 |
| SBS | Chr3 | 26675691 | C-T | Homo | ||
| SBS | Chr3 | 26675691 | C-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 6023055 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 6899654 | C-T | HET | ||
| SBS | Chr3 | 6899654 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 1234456 | C-T | HET | ||
| SBS | Chr4 | 1234456 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 12421217 | C-T | Homo | ||
| SBS | Chr4 | 12421217 | C-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 12679842 | G-A | HET | ||
| SBS | Chr4 | 22468058 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g37790 |
| SBS | Chr4 | 23624265 | G-T | HET | INTERGENIC | |
| SBS | Chr4 | 2377529 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g04930 |
| SBS | Chr4 | 463253 | T-C | HET | ||
| SBS | Chr4 | 463253 | T-C | HET | INTERGENIC | |
| SBS | Chr5 | 14045367 | G-A | HET | ||
| SBS | Chr5 | 14749903 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os05g25390 |
| SBS | Chr5 | 16307454 | T-C | HET | ||
| SBS | Chr5 | 16307454 | T-C | HET | INTRON | |
| SBS | Chr5 | 16328490 | G-A | HET | ||
| SBS | Chr5 | 16328490 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 16328495 | G-A | HET | ||
| SBS | Chr5 | 16328495 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 16328499 | G-A | HET | ||
| SBS | Chr5 | 16328499 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 17504280 | C-T | HET | ||
| SBS | Chr5 | 25173778 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 5773931 | T-A | HET | ||
| SBS | Chr5 | 5864684 | G-A | HET | ||
| SBS | Chr5 | 5864684 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 8228097 | C-T | HET | ||
| SBS | Chr5 | 8662127 | G-A | HET | ||
| SBS | Chr5 | 8662127 | G-A | HET | INTRON | |
| SBS | Chr5 | 8724279 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os05g15430 |
| SBS | Chr5 | 9584360 | T-A | HET | ||
| SBS | Chr6 | 15165863 | C-T | HET | ||
| SBS | Chr6 | 21553626 | T-A | Homo | ||
| SBS | Chr6 | 21553626 | T-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 24628735 | A-T | HET | ||
| SBS | Chr6 | 24628735 | A-T | HET | INTERGENIC | |
| SBS | Chr7 | 1789930 | A-G | HET | INTRON | |
| SBS | Chr7 | 19044128 | T-C | HET | INTERGENIC | |
| SBS | Chr7 | 19156138 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 24597819 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g41090 |
| SBS | Chr7 | 8404042 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 16989257 | G-A | HET | ||
| SBS | Chr8 | 16989257 | G-A | HET | INTRON | |
| SBS | Chr8 | 21433171 | T-C | HET | ||
| SBS | Chr8 | 27712017 | A-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os08g44015 |
| SBS | Chr8 | 27712017 | A-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os08g44015 |
| SBS | Chr8 | 28315571 | T-A | HET | ||
| SBS | Chr8 | 3132034 | C-T | HET | ||
| SBS | Chr8 | 3132034 | C-T | HET | UTR_5_PRIME | |
| SBS | Chr8 | 4540191 | T-A | HET | ||
| SBS | Chr8 | 5276209 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os08g09110 |
| SBS | Chr8 | 8807498 | C-T | HOMO | INTERGENIC | |
| SBS | Chr9 | 14324504 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 21749431 | C-T | HET | ||
| SBS | Chr9 | 21749431 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 849024 | T-C | HET | ||
| SBS | Chr9 | 849024 | T-C | HET | INTERGENIC |
Deletions: 45
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr7 | 641171 | 641175 | 4 | |
| Deletion | Chr2 | 886001 | 894000 | 7999 | 3 |
| Deletion | Chr9 | 2206044 | 2206073 | 29 | LOC_Os09g04190 |
| Deletion | Chr12 | 2335196 | 2335205 | 9 | |
| Deletion | Chr6 | 3688367 | 3688373 | 6 | 2 |
| Deletion | Chr12 | 3714341 | 3714346 | 5 | |
| Deletion | Chr6 | 4356001 | 4366000 | 9999 | LOC_Os06g08884 |
| Deletion | Chr5 | 4517990 | 4517996 | 6 | |
| Deletion | Chr10 | 4808925 | 4808933 | 8 | |
| Deletion | Chr11 | 5212632 | 5212636 | 4 | LOC_Os11g09760 |
| Deletion | Chr11 | 5566720 | 5566721 | 1 | |
| Deletion | Chr9 | 6106051 | 6106052 | 1 | |
| Deletion | Chr4 | 6198369 | 6198372 | 3 | LOC_Os04g11350 |
| Deletion | Chr12 | 6410317 | 6410318 | 1 | |
| Deletion | Chr5 | 6779001 | 7262000 | 482999 | 72 |
| Deletion | Chr5 | 6790001 | 7345000 | 554999 | 87 |
| Deletion | Chr5 | 7267001 | 7347000 | 79999 | 14 |
| Deletion | Chr8 | 7489187 | 7489215 | 28 | LOC_Os08g12680 |
| Deletion | Chr8 | 8537079 | 8537080 | 1 | |
| Deletion | Chr3 | 10416501 | 10416508 | 7 | 2 |
| Deletion | Chr11 | 11802817 | 11802819 | 2 | |
| Deletion | Chr11 | 12300301 | 12300465 | 164 | |
| Deletion | Chr11 | 13233641 | 13233643 | 2 | LOC_Os11g23010 |
| Deletion | Chr11 | 14094682 | 14094693 | 11 | |
| Deletion | Chr10 | 14212817 | 14212818 | 1 | |
| Deletion | Chr4 | 14661769 | 14661772 | 3 | 2 |
| Deletion | Chr9 | 15626546 | 15626554 | 8 | 2 |
| Deletion | Chr8 | 15885406 | 15885410 | 4 | 2 |
| Deletion | Chr5 | 16249001 | 16273000 | 23999 | 2 |
| Deletion | Chr5 | 16254001 | 16273000 | 18999 | 2 |
| Deletion | Chr11 | 16566655 | 16566666 | 11 | |
| Deletion | Chr8 | 16576616 | 16576617 | 1 | |
| Deletion | Chr11 | 16687526 | 16687527 | 1 | LOC_Os11g28790 |
| Deletion | Chr11 | 17941434 | 17941439 | 5 | |
| Deletion | Chr5 | 18706226 | 18706251 | 25 | |
| Deletion | Chr10 | 19721746 | 19721747 | 1 | |
| Deletion | Chr9 | 20678867 | 20678878 | 11 | |
| Deletion | Chr9 | 21196614 | 21196629 | 15 | |
| Deletion | Chr12 | 21733136 | 21733145 | 9 | 2 |
| Deletion | Chr11 | 23873974 | 23873976 | 2 | |
| Deletion | Chr6 | 25819688 | 25819689 | 1 | 2 |
| Deletion | Chr7 | 25999081 | 25999082 | 1 | 2 |
| Deletion | Chr1 | 35562581 | 35562582 | 1 | 2 |
| Deletion | Chr1 | 42526924 | 42526927 | 3 | LOC_Os01g73370 |
| Deletion | Chr1 | 42586031 | 42586037 | 6 |
Insertions: 10
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 21801636 | 21801636 | 1 | |
| Insertion | Chr12 | 11575371 | 11575371 | 1 | |
| Insertion | Chr12 | 17467227 | 17467228 | 2 | |
| Insertion | Chr2 | 15911341 | 15911342 | 2 | 2 |
| Insertion | Chr2 | 15911341 | 15911342 | 2 | 2 |
| Insertion | Chr2 | 19774202 | 19774202 | 1 | |
| Insertion | Chr6 | 22480986 | 22480986 | 1 | |
| Insertion | Chr8 | 1330572 | 1330573 | 2 | |
| Insertion | Chr8 | 8918680 | 8918689 | 10 | 2 |
| Insertion | Chr8 | 8918680 | 8918687 | 8 | 2 |
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr5 | 16251355 | 16296539 | 2 |
| Inversion | Chr5 | 16271253 | 16296627 | 2 |
| Inversion | Chr2 | 28127867 | 28128197 | LOC_Os02g46150 |
| Inversion | Chr3 | 33882604 | 34273852 |
Translocations: 8
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 4699954 | Chr7 | 27254265 | |
| Translocation | Chr10 | 5149237 | Chr4 | 3713485 | |
| Translocation | Chr7 | 9320330 | Chr4 | 3213283 | LOC_Os07g15990 |
| Translocation | Chr6 | 10375738 | Chr3 | 22587351 | |
| Translocation | Chr7 | 11905744 | Chr4 | 16178237 | LOC_Os04g27380 |
| Translocation | Chr8 | 14880098 | Chr3 | 23141741 | |
| Translocation | Chr12 | 16446401 | Chr7 | 11777249 | LOC_Os12g27900 |
| Translocation | Chr12 | 24332920 | Chr3 | 20216552 |
