Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2708-S-1 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2708-S-1 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 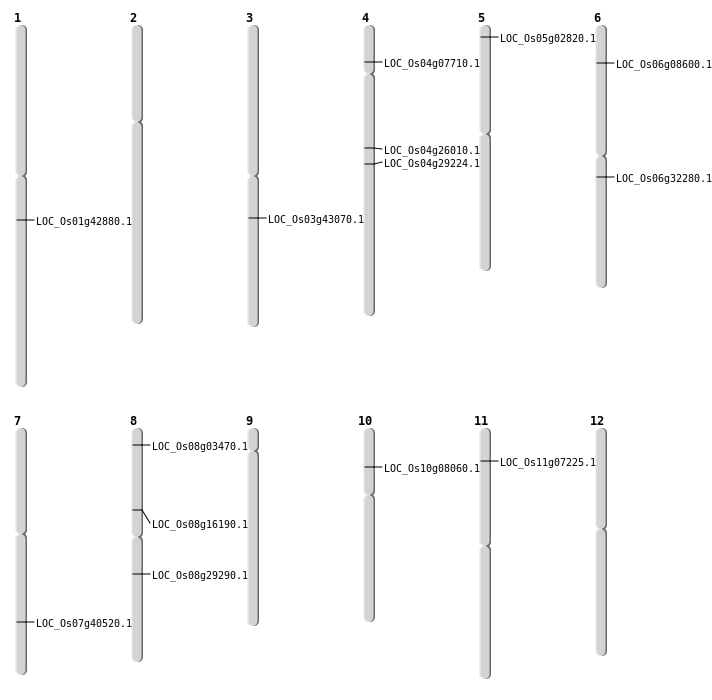
|
| Variant Information |
|---|
Single base substitutions: 52
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 10777531 | G-A | HET | INTRON | |
| SBS | Chr1 | 17114564 | T-C | HET | INTERGENIC | |
| SBS | Chr1 | 24398279 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g42880 |
| SBS | Chr1 | 32743063 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 33799752 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 43024685 | A-T | HOMO | INTERGENIC | |
| SBS | Chr10 | 19957711 | A-G | HET | INTERGENIC | |
| SBS | Chr10 | 4369908 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os10g08060 |
| SBS | Chr10 | 8545410 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr10 | 8660603 | A-G | HET | INTERGENIC | |
| SBS | Chr11 | 2293021 | T-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 23201344 | A-T | HOMO | INTERGENIC | |
| SBS | Chr11 | 23201345 | A-T | HOMO | INTERGENIC | |
| SBS | Chr11 | 3654451 | A-G | HOMO | INTERGENIC | |
| SBS | Chr11 | 9383139 | A-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr11 | 9478083 | G-A | HET | INTRON | |
| SBS | Chr12 | 10792621 | A-T | HET | INTERGENIC | |
| SBS | Chr12 | 12474125 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 4277653 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 18024864 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 28182669 | C-G | HOMO | INTERGENIC | |
| SBS | Chr3 | 12228120 | A-T | HET | INTERGENIC | |
| SBS | Chr3 | 15341852 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 21552725 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 28274552 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr3 | 34238255 | A-C | HET | UTR_3_PRIME | |
| SBS | Chr4 | 12126014 | G-T | HET | INTERGENIC | |
| SBS | Chr4 | 13651939 | T-A | HET | INTRON | |
| SBS | Chr4 | 1382552 | G-A | HET | INTRON | |
| SBS | Chr4 | 16935597 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 1944 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 6110329 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 1016298 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os05g02820 |
| SBS | Chr5 | 10238432 | C-A | HET | INTERGENIC | |
| SBS | Chr5 | 21652238 | T-C | HET | INTERGENIC | |
| SBS | Chr5 | 6878934 | G-A | HET | INTRON | |
| SBS | Chr6 | 10442619 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 13061222 | A-C | HET | INTERGENIC | |
| SBS | Chr6 | 16283453 | T-A | HET | INTERGENIC | |
| SBS | Chr6 | 16283454 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 19732995 | T-A | HET | INTERGENIC | |
| SBS | Chr7 | 11327689 | T-A | HET | INTRON | |
| SBS | Chr7 | 16536956 | A-T | HET | INTERGENIC | |
| SBS | Chr7 | 24274756 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os07g40520 |
| SBS | Chr7 | 7899148 | C-T | HOMO | INTERGENIC | |
| SBS | Chr8 | 17085853 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 9872965 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os08g16190 |
| SBS | Chr9 | 1133566 | G-T | HET | INTRON | |
| SBS | Chr9 | 12661658 | T-A | HET | INTERGENIC | |
| SBS | Chr9 | 15101564 | G-C | HET | INTERGENIC | |
| SBS | Chr9 | 4502118 | T-C | HET | INTERGENIC | |
| SBS | Chr9 | 4533826 | A-G | HET | INTERGENIC |
Deletions: 21
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr10 | 528110 | 528111 | 1 | |
| Deletion | Chr8 | 1629903 | 1629912 | 9 | LOC_Os08g03470 |
| Deletion | Chr4 | 4095918 | 4095921 | 3 | LOC_Os04g07710 |
| Deletion | Chr1 | 12162122 | 12162150 | 28 | |
| Deletion | Chr11 | 15245050 | 15245051 | 1 | |
| Deletion | Chr12 | 15532350 | 15532351 | 1 | |
| Deletion | Chr4 | 17329863 | 17329864 | 1 | LOC_Os04g29224 |
| Deletion | Chr8 | 17973215 | 17973216 | 1 | LOC_Os08g29290 |
| Deletion | Chr6 | 18794602 | 18794626 | 24 | LOC_Os06g32280 |
| Deletion | Chr6 | 20859265 | 20859268 | 3 | |
| Deletion | Chr1 | 21759427 | 21759428 | 1 | |
| Deletion | Chr6 | 22482944 | 22482954 | 10 | |
| Deletion | Chr12 | 24258687 | 24258697 | 10 | |
| Deletion | Chr8 | 24956181 | 24956188 | 7 | |
| Deletion | Chr6 | 25923832 | 25923842 | 10 | |
| Deletion | Chr7 | 27362691 | 27362698 | 7 | |
| Deletion | Chr11 | 28345609 | 28345613 | 4 | |
| Deletion | Chr4 | 29597604 | 29597605 | 1 | |
| Deletion | Chr3 | 29966319 | 29966320 | 1 | |
| Deletion | Chr2 | 30643102 | 30643104 | 2 | |
| Deletion | Chr3 | 36032971 | 36032974 | 3 |
Insertions: 14
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr10 | 17036647 | 17036674 | 28 | |
| Insertion | Chr10 | 3458004 | 3458042 | 39 | |
| Insertion | Chr11 | 23784729 | 23784729 | 1 | |
| Insertion | Chr11 | 27074008 | 27074008 | 1 | |
| Insertion | Chr2 | 1587011 | 1587012 | 2 | |
| Insertion | Chr2 | 22207158 | 22207159 | 2 | |
| Insertion | Chr2 | 34283517 | 34283517 | 1 | |
| Insertion | Chr4 | 15138019 | 15138019 | 1 | LOC_Os04g26010 |
| Insertion | Chr6 | 18652730 | 18652730 | 1 | |
| Insertion | Chr7 | 11392750 | 11392750 | 1 | |
| Insertion | Chr7 | 19605296 | 19605298 | 3 | |
| Insertion | Chr7 | 20462920 | 20462957 | 38 | |
| Insertion | Chr8 | 24612150 | 24612150 | 1 | |
| Insertion | Chr9 | 13436046 | 13436065 | 20 |
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr11 | 3629814 | 3656456 | LOC_Os11g07225 |
| Inversion | Chr11 | 3629818 | 3656463 | LOC_Os11g07225 |
| Inversion | Chr10 | 5787856 | 6277082 |
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr7 | 623317 | Chr3 | 22576444 | |
| Translocation | Chr8 | 8367591 | Chr2 | 16568341 | |
| Translocation | Chr9 | 16167563 | Chr6 | 4276869 | LOC_Os06g08600 |
| Translocation | Chr4 | 22819421 | Chr3 | 24030531 | LOC_Os03g43070 |
| Translocation | Chr11 | 28275633 | Chr4 | 22819310 |
