Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2709-S-1 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2709-S-1 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 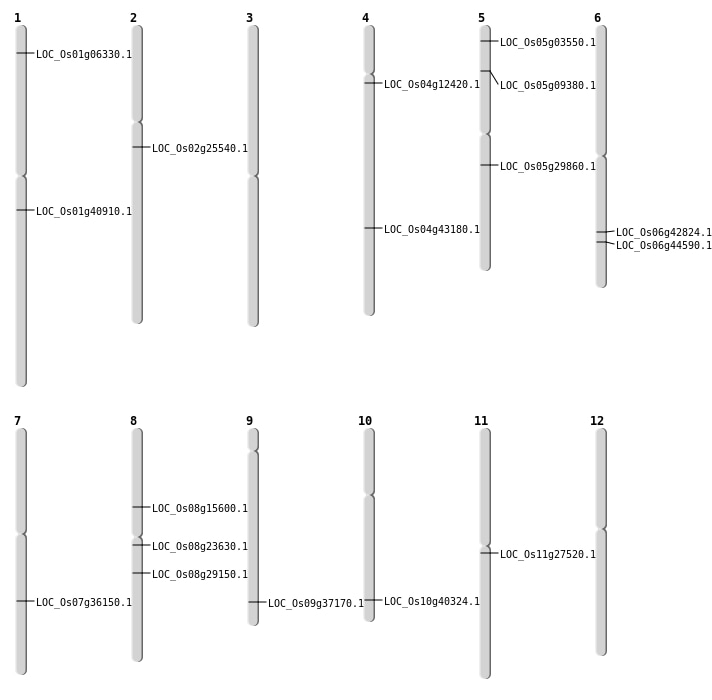
|
| Variant Information |
|---|
Single base substitutions: 80
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 16429382 | G-T | HET | INTRON | |
| SBS | Chr1 | 17287297 | G-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 19014604 | T-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 20235219 | C-G | HET | INTRON | |
| SBS | Chr1 | 21306639 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr1 | 22625669 | T-A | HET | INTRON | |
| SBS | Chr1 | 23139586 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g40910 |
| SBS | Chr1 | 28637786 | C-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 32885325 | T-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 41752625 | T-A | HET | INTERGENIC | |
| SBS | Chr1 | 41752633 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 679274 | C-T | HET | INTRON | |
| SBS | Chr10 | 13397231 | C-A | HET | INTRON | |
| SBS | Chr10 | 18637152 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 23073199 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 4022738 | A-T | HET | INTERGENIC | |
| SBS | Chr10 | 920617 | C-G | HET | INTERGENIC | |
| SBS | Chr10 | 9571037 | A-G | HET | INTERGENIC | |
| SBS | Chr11 | 15834268 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os11g27520 |
| SBS | Chr11 | 23691196 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 26448238 | A-T | HET | INTERGENIC | |
| SBS | Chr11 | 27216864 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 16057394 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 19067694 | C-A | HET | INTERGENIC | |
| SBS | Chr12 | 19067696 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 25770138 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g41650 |
| SBS | Chr12 | 4669337 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 4930183 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 6912079 | C-T | HET | INTRON | |
| SBS | Chr12 | 8552992 | G-C | HET | INTERGENIC | |
| SBS | Chr2 | 10533453 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 21724644 | C-G | HET | INTRON | |
| SBS | Chr2 | 34448297 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 11053618 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 14063611 | A-G | HET | INTRON | |
| SBS | Chr3 | 14748060 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 20011166 | T-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 23819291 | G-A | HET | INTRON | |
| SBS | Chr3 | 32911488 | G-T | HET | UTR_3_PRIME | |
| SBS | Chr3 | 6470887 | A-G | HET | INTRON | |
| SBS | Chr4 | 21584925 | C-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr4 | 21585263 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr4 | 24094223 | T-A | HOMO | INTRON | |
| SBS | Chr4 | 25559052 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g43180 |
| SBS | Chr4 | 26486046 | T-G | HET | INTRON | |
| SBS | Chr4 | 27920078 | A-T | HOMO | INTRON | |
| SBS | Chr4 | 27975544 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 30230349 | C-A | HET | START_GAINED | |
| SBS | Chr4 | 3553622 | A-C | HET | INTERGENIC | |
| SBS | Chr4 | 6859011 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g12420 |
| SBS | Chr4 | 7000 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr4 | 8194449 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 1502007 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g03550 |
| SBS | Chr5 | 23303591 | G-T | HET | INTERGENIC | |
| SBS | Chr5 | 23303592 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 25904350 | A-G | HET | INTERGENIC | |
| SBS | Chr5 | 29795630 | A-G | HET | INTERGENIC | |
| SBS | Chr5 | 5269325 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g09380 |
| SBS | Chr5 | 7778310 | G-A | HET | INTRON | |
| SBS | Chr6 | 10664073 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 3681366 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 10010531 | C-G | HET | INTERGENIC | |
| SBS | Chr7 | 10471460 | G-C | HET | INTERGENIC | |
| SBS | Chr7 | 11578448 | C-A | HET | INTERGENIC | |
| SBS | Chr7 | 11933528 | G-C | HET | INTERGENIC | |
| SBS | Chr7 | 21595020 | T-A | HET | STOP_GAINED | LOC_Os07g36150 |
| SBS | Chr7 | 28386356 | T-C | HET | INTERGENIC | |
| SBS | Chr8 | 10953767 | T-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr8 | 13069668 | A-T | HET | INTERGENIC | |
| SBS | Chr8 | 14304167 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g23630 |
| SBS | Chr8 | 17845289 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g29150 |
| SBS | Chr8 | 21933266 | C-A | HET | INTERGENIC | |
| SBS | Chr8 | 23227320 | G-A | HET | INTRON | |
| SBS | Chr8 | 24707443 | G-T | HET | INTERGENIC | |
| SBS | Chr8 | 4366812 | A-G | HET | INTERGENIC | |
| SBS | Chr8 | 6183155 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 8638721 | A-G | HET | INTERGENIC | |
| SBS | Chr9 | 12273396 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 17071614 | T-C | HOMO | INTERGENIC | |
| SBS | Chr9 | 603754 | T-C | HET | INTERGENIC |
Deletions: 20
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr9 | 2449714 | 2449715 | 1 | |
| Deletion | Chr8 | 2710572 | 2710573 | 1 | |
| Deletion | Chr9 | 2820208 | 2820209 | 1 | |
| Deletion | Chr3 | 3972141 | 3972142 | 1 | |
| Deletion | Chr3 | 4131267 | 4131275 | 8 | |
| Deletion | Chr10 | 5084382 | 5084383 | 1 | |
| Deletion | Chr1 | 5229966 | 5229967 | 1 | |
| Deletion | Chr2 | 10185616 | 10185628 | 12 | |
| Deletion | Chr5 | 10667149 | 10667152 | 3 | |
| Deletion | Chr1 | 12456551 | 12456552 | 1 | |
| Deletion | Chr10 | 13117777 | 13117779 | 2 | |
| Deletion | Chr10 | 14496026 | 14496031 | 5 | |
| Deletion | Chr5 | 17762176 | 17762177 | 1 | |
| Deletion | Chr9 | 21442116 | 21442151 | 35 | LOC_Os09g37170 |
| Deletion | Chr2 | 25167600 | 25167601 | 1 | |
| Deletion | Chr1 | 26555324 | 26555325 | 1 | |
| Deletion | Chr8 | 27820285 | 27820287 | 2 | |
| Deletion | Chr7 | 28685699 | 28685700 | 1 | |
| Deletion | Chr3 | 35617386 | 35617393 | 7 | |
| Deletion | Chr1 | 42908486 | 42908488 | 2 |
Insertions: 14
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 38467392 | 38467392 | 1 | |
| Insertion | Chr11 | 27146858 | 27146885 | 28 | |
| Insertion | Chr12 | 25845091 | 25845091 | 1 | |
| Insertion | Chr2 | 14922677 | 14922677 | 1 | LOC_Os02g25540 |
| Insertion | Chr3 | 21803720 | 21803720 | 1 | |
| Insertion | Chr4 | 26501103 | 26501104 | 2 | |
| Insertion | Chr4 | 26734141 | 26734142 | 2 | |
| Insertion | Chr6 | 11000810 | 11000810 | 1 | |
| Insertion | Chr6 | 26926156 | 26926156 | 1 | LOC_Os06g44590 |
| Insertion | Chr7 | 27289833 | 27289834 | 2 | |
| Insertion | Chr7 | 7456695 | 7456695 | 1 | |
| Insertion | Chr8 | 17507783 | 17507825 | 43 | |
| Insertion | Chr8 | 22787352 | 22787352 | 1 | |
| Insertion | Chr8 | 9482107 | 9482107 | 1 | LOC_Os08g15600 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr1 | 32659264 | 33203810 | |
| Inversion | Chr1 | 32659277 | 33203810 |
Translocations: 11
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr8 | 5248589 | Chr1 | 39255030 | |
| Translocation | Chr8 | 5252904 | Chr1 | 39099944 | |
| Translocation | Chr9 | 7749089 | Chr5 | 15268435 | |
| Translocation | Chr9 | 7749098 | Chr5 | 15268863 | |
| Translocation | Chr8 | 16734275 | Chr6 | 17008860 | |
| Translocation | Chr5 | 17279091 | Chr2 | 11136431 | LOC_Os05g29860 |
| Translocation | Chr5 | 17279093 | Chr2 | 11135897 | LOC_Os05g29860 |
| Translocation | Chr10 | 21612546 | Chr6 | 25743065 | 2 |
| Translocation | Chr11 | 22094146 | Chr8 | 7405061 | |
| Translocation | Chr2 | 22548586 | Chr1 | 3012122 | LOC_Os01g06330 |
| Translocation | Chr11 | 22753380 | Chr2 | 5999994 |
