Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2714-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2714-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 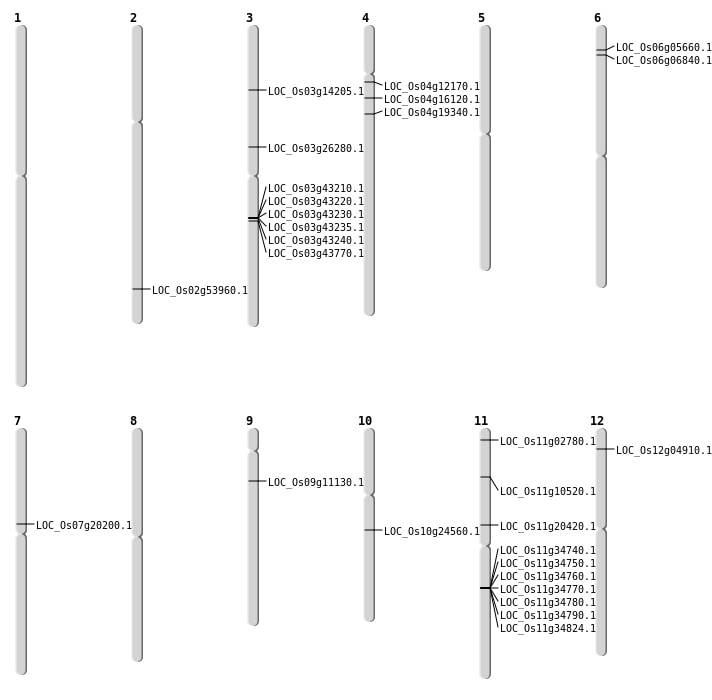
|
| Variant Information |
|---|
Single base substitutions: 46
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 11559019 | C-G | HOMO | INTERGENIC | |
| SBS | Chr1 | 26826808 | G-T | HET | INTERGENIC | |
| SBS | Chr1 | 34522818 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 42073721 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 12604490 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g24560 |
| SBS | Chr10 | 21909551 | G-A | HET | INTRON | |
| SBS | Chr10 | 5080053 | A-G | HET | INTERGENIC | |
| SBS | Chr11 | 11815349 | C-T | HET | STOP_GAINED | LOC_Os11g20420 |
| SBS | Chr11 | 12667050 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 13327173 | G-A | HET | INTRON | |
| SBS | Chr11 | 926269 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g02780 |
| SBS | Chr12 | 16158814 | C-G | HET | INTERGENIC | |
| SBS | Chr2 | 10724682 | C-G | HET | INTERGENIC | |
| SBS | Chr2 | 14329557 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 20537786 | G-A | HET | INTRON | |
| SBS | Chr2 | 20895932 | T-C | HET | INTERGENIC | |
| SBS | Chr2 | 22419181 | G-C | HET | INTERGENIC | |
| SBS | Chr2 | 33038448 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os02g53960 |
| SBS | Chr3 | 10937982 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 15029788 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g26280 |
| SBS | Chr3 | 16317897 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 28083227 | G-C | HET | INTERGENIC | |
| SBS | Chr3 | 30377951 | C-G | HET | INTERGENIC | |
| SBS | Chr3 | 32260104 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 14846979 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 1587853 | A-C | HET | INTERGENIC | |
| SBS | Chr4 | 21923782 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr4 | 23219583 | T-G | HET | INTERGENIC | |
| SBS | Chr4 | 24314201 | A-G | HET | INTRON | |
| SBS | Chr4 | 6706688 | A-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os04g12170 |
| SBS | Chr4 | 8751430 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os04g16120 |
| SBS | Chr5 | 10705316 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 10705320 | T-G | HET | INTERGENIC | |
| SBS | Chr5 | 25687327 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 8589700 | T-C | HET | INTERGENIC | |
| SBS | Chr6 | 15307702 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 2560925 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os06g05660 |
| SBS | Chr6 | 26105902 | C-G | HOMO | INTRON | |
| SBS | Chr6 | 994243 | T-C | HOMO | INTERGENIC | |
| SBS | Chr7 | 26878267 | A-C | HET | INTERGENIC | |
| SBS | Chr7 | 27216921 | G-A | HET | INTRON | |
| SBS | Chr7 | 27216922 | G-A | HET | INTRON | |
| SBS | Chr8 | 15720756 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 2606293 | G-T | HET | INTERGENIC | |
| SBS | Chr8 | 8303236 | C-A | HOMO | INTRON | |
| SBS | Chr9 | 10852941 | A-G | HOMO | UTR_5_PRIME |
Deletions: 21
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr9 | 6168442 | 6168448 | 6 | LOC_Os09g11130 |
| Deletion | Chr12 | 8260618 | 8260626 | 8 | |
| Deletion | Chr4 | 8850262 | 8850263 | 1 | |
| Deletion | Chr7 | 9666603 | 9666604 | 1 | |
| Deletion | Chr5 | 10096998 | 10097000 | 2 | |
| Deletion | Chr4 | 10761252 | 10761253 | 1 | LOC_Os04g19340 |
| Deletion | Chr9 | 11035779 | 11035791 | 12 | |
| Deletion | Chr7 | 11834401 | 11834402 | 1 | |
| Deletion | Chr9 | 13377384 | 13377385 | 1 | |
| Deletion | Chr6 | 14476682 | 14476686 | 4 | |
| Deletion | Chr10 | 14863025 | 14863026 | 1 | |
| Deletion | Chr12 | 15683807 | 15683808 | 1 | |
| Deletion | Chr8 | 16945564 | 16945566 | 2 | |
| Deletion | Chr11 | 20352001 | 20385000 | 32999 | 7 |
| Deletion | Chr7 | 21788771 | 21788785 | 14 | |
| Deletion | Chr3 | 24090001 | 24112000 | 21999 | 6 |
| Deletion | Chr4 | 25220379 | 25220441 | 62 | |
| Deletion | Chr8 | 27229104 | 27229124 | 20 | |
| Deletion | Chr5 | 29855358 | 29855712 | 354 | |
| Deletion | Chr6 | 30712942 | 30712943 | 1 | |
| Deletion | Chr2 | 32415787 | 32415791 | 4 |
Insertions: 8
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 23210113 | 23210116 | 4 | |
| Insertion | Chr11 | 13568674 | 13568674 | 1 | |
| Insertion | Chr12 | 24097878 | 24097879 | 2 | |
| Insertion | Chr3 | 7706778 | 7706778 | 1 | LOC_Os03g14205 |
| Insertion | Chr4 | 4953974 | 4953974 | 1 | |
| Insertion | Chr6 | 15971205 | 15971212 | 8 | |
| Insertion | Chr7 | 9296576 | 9296577 | 2 | |
| Insertion | Chr8 | 7269088 | 7269088 | 1 |
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr11 | 19287995 | 19561891 | |
| Inversion | Chr11 | 19443579 | 19460301 | |
| Inversion | Chr11 | 19561882 | 19580201 |
Translocations: 16
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 2109749 | Chr9 | 9468868 | LOC_Os12g04910 |
| Translocation | Chr9 | 2318979 | Chr2 | 16818229 | |
| Translocation | Chr7 | 2877918 | Chr6 | 26188660 | |
| Translocation | Chr5 | 3543878 | Chr3 | 13563773 | |
| Translocation | Chr7 | 4604537 | Chr1 | 7703254 | |
| Translocation | Chr11 | 5756374 | Chr1 | 31898866 | LOC_Os11g10520 |
| Translocation | Chr11 | 5756424 | Chr1 | 31897578 | LOC_Os11g10520 |
| Translocation | Chr11 | 6603603 | Chr3 | 664990 | |
| Translocation | Chr10 | 6833638 | Chr6 | 18384660 | |
| Translocation | Chr9 | 10373260 | Chr2 | 17573565 | |
| Translocation | Chr7 | 11660419 | Chr6 | 3229970 | 2 |
| Translocation | Chr11 | 15465476 | Chr1 | 41568841 | |
| Translocation | Chr12 | 18566350 | Chr3 | 15160099 | |
| Translocation | Chr8 | 20336255 | Chr4 | 2842147 | |
| Translocation | Chr6 | 20343644 | Chr4 | 13171073 | |
| Translocation | Chr4 | 35497981 | Chr1 | 42743842 |
