Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2715-S [Download] |
| Generation | M2-SOS |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2715-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 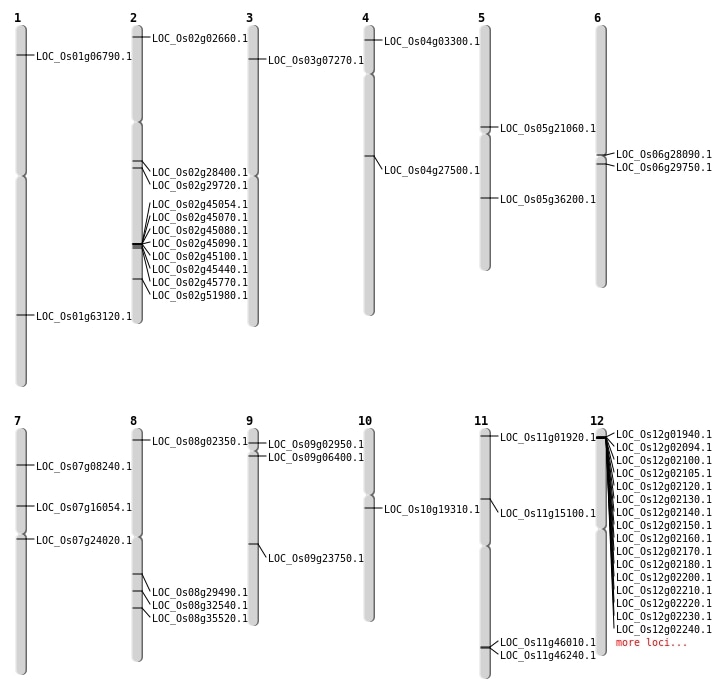
|
| Variant Information |
|---|
Single base substitutions: 105
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 16494888 | A-T | HET | ||
| SBS | Chr1 | 16494888 | A-T | HET | INTERGENIC | |
| SBS | Chr1 | 17987967 | G-T | HET | INTERGENIC | |
| SBS | Chr1 | 24625957 | A-G | HET | ||
| SBS | Chr1 | 28967030 | C-T | HET | ||
| SBS | Chr1 | 36577128 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g63120 |
| SBS | Chr1 | 41169498 | G-A | HET | ||
| SBS | Chr1 | 41169498 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 7819929 | C-A | HET | ||
| SBS | Chr10 | 1353650 | G-A | Homo | ||
| SBS | Chr10 | 1353650 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr10 | 142035 | C-A | HET | ||
| SBS | Chr10 | 20488813 | G-C | HET | ||
| SBS | Chr10 | 20488813 | G-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr10 | 3914015 | A-G | HET | ||
| SBS | Chr10 | 670843 | C-T | HET | ||
| SBS | Chr10 | 9679924 | C-T | HET | ||
| SBS | Chr10 | 9841160 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os10g19310 |
| SBS | Chr11 | 23859421 | G-A | HET | ||
| SBS | Chr11 | 27332789 | C-A | Homo | ||
| SBS | Chr11 | 27332789 | C-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 27843437 | C-G | HET | ||
| SBS | Chr11 | 28003589 | T-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os11g46240 |
| SBS | Chr11 | 28003589 | T-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os11g46240 |
| SBS | Chr11 | 3134868 | C-T | HET | ||
| SBS | Chr11 | 493845 | C-T | HET | STOP_GAINED | LOC_Os11g01920 |
| SBS | Chr11 | 765546 | A-G | HET | ||
| SBS | Chr11 | 8498812 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os11g15100 |
| SBS | Chr11 | 8498812 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os11g15100 |
| SBS | Chr12 | 18084217 | G-T | HET | INTERGENIC | |
| SBS | Chr2 | 1111626 | C-G | HET | ||
| SBS | Chr2 | 15799469 | C-T | HET | ||
| SBS | Chr2 | 15799469 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 17676786 | G-C | HOMO | NON_SYNONYMOUS_CODING | LOC_Os02g29720 |
| SBS | Chr2 | 17676786 | G-C | Homo | NON_SYNONYMOUS_CODING | LOC_Os02g29720 |
| SBS | Chr2 | 26126958 | T-A | HET | UTR_3_PRIME | |
| SBS | Chr2 | 30620881 | G-T | HET | INTERGENIC | |
| SBS | Chr2 | 31827691 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g51980 |
| SBS | Chr2 | 8759149 | G-T | HET | INTERGENIC | |
| SBS | Chr2 | 981648 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os02g02660 |
| SBS | Chr3 | 12912504 | C-A | HET | ||
| SBS | Chr3 | 16709920 | A-T | HET | INTERGENIC | |
| SBS | Chr3 | 24126108 | T-C | HET | INTRON | |
| SBS | Chr3 | 35006865 | T-C | HET | ||
| SBS | Chr3 | 35006865 | T-C | HET | INTRON | |
| SBS | Chr3 | 35498280 | G-T | HET | ||
| SBS | Chr3 | 35498280 | G-T | HET | INTERGENIC | |
| SBS | Chr3 | 35498293 | T-A | HET | ||
| SBS | Chr3 | 35498293 | T-A | HET | INTERGENIC | |
| SBS | Chr3 | 775519 | T-G | HET | ||
| SBS | Chr3 | 775519 | T-G | HET | INTRON | |
| SBS | Chr4 | 13556012 | G-A | HET | ||
| SBS | Chr4 | 1406427 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g03300 |
| SBS | Chr4 | 14258771 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 16859431 | C-A | HET | ||
| SBS | Chr4 | 19787876 | G-A | HET | ||
| SBS | Chr4 | 19787876 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 23452239 | G-A | HET | ||
| SBS | Chr4 | 23720659 | G-T | HET | INTRON | |
| SBS | Chr4 | 31052968 | A-G | HET | INTERGENIC | |
| SBS | Chr4 | 33352948 | G-A | HET | ||
| SBS | Chr4 | 34548856 | T-C | HET | ||
| SBS | Chr4 | 9654766 | A-C | HET | ||
| SBS | Chr4 | 9654766 | A-C | HET | INTERGENIC | |
| SBS | Chr5 | 12387736 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g21060 |
| SBS | Chr5 | 17836577 | G-A | HET | ||
| SBS | Chr5 | 21468185 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os05g36200 |
| SBS | Chr5 | 22372783 | G-A | HET | ||
| SBS | Chr5 | 22372783 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 26519433 | A-T | HET | INTERGENIC | |
| SBS | Chr5 | 29642874 | C-T | HET | ||
| SBS | Chr5 | 29642874 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 4433346 | C-T | Homo | ||
| SBS | Chr5 | 4433346 | C-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 12353668 | T-A | HET | INTRON | |
| SBS | Chr6 | 15955360 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os06g28090 |
| SBS | Chr6 | 17047476 | C-T | HET | ||
| SBS | Chr6 | 6137972 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 14912354 | T-A | HET | INTERGENIC | |
| SBS | Chr7 | 15550437 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 18519318 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 26012668 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 4200893 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g08240 |
| SBS | Chr7 | 4967600 | G-T | HET | ||
| SBS | Chr7 | 4967600 | G-T | HET | INTERGENIC | |
| SBS | Chr7 | 502927 | A-T | HET | ||
| SBS | Chr7 | 502927 | A-T | HET | INTERGENIC | |
| SBS | Chr8 | 16298191 | A-T | HET | INTERGENIC | |
| SBS | Chr8 | 16840083 | T-C | HET | INTERGENIC | |
| SBS | Chr8 | 16973994 | A-T | HET | INTERGENIC | |
| SBS | Chr8 | 18073683 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g29490 |
| SBS | Chr8 | 21445582 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 23345956 | G-T | HET | INTERGENIC | |
| SBS | Chr8 | 2831370 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 4898554 | T-G | Homo | ||
| SBS | Chr8 | 4898554 | T-G | HOMO | INTERGENIC | |
| SBS | Chr8 | 920546 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os08g02350 |
| SBS | Chr9 | 14126947 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os09g23750 |
| SBS | Chr9 | 1649877 | G-T | Homo | ||
| SBS | Chr9 | 1649877 | G-T | HOMO | INTERGENIC | |
| SBS | Chr9 | 18820978 | A-T | HET | ||
| SBS | Chr9 | 18820978 | A-T | HET | INTERGENIC | |
| SBS | Chr9 | 21922234 | G-A | HET | ||
| SBS | Chr9 | 245584 | T-G | Homo | ||
| SBS | Chr9 | 245584 | T-G | HOMO | UTR_3_PRIME |
Deletions: 42
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr12 | 628001 | 750000 | 121999 | 23 |
| Deletion | Chr12 | 629001 | 747000 | 117999 | 24 |
| Deletion | Chr10 | 3478370 | 3478371 | 1 | |
| Deletion | Chr5 | 4044435 | 4044442 | 7 | 2 |
| Deletion | Chr9 | 5503737 | 5503739 | 2 | |
| Deletion | Chr8 | 6386098 | 6386099 | 1 | 2 |
| Deletion | Chr8 | 6443606 | 6443634 | 28 | |
| Deletion | Chr2 | 6893754 | 6893755 | 1 | |
| Deletion | Chr1 | 7861415 | 7861419 | 4 | |
| Deletion | Chr12 | 8315161 | 8315162 | 1 | |
| Deletion | Chr7 | 8429714 | 8429715 | 1 | |
| Deletion | Chr9 | 8845257 | 8845258 | 1 | |
| Deletion | Chr7 | 9359689 | 9359690 | 1 | LOC_Os07g16054 |
| Deletion | Chr7 | 9793142 | 9793143 | 1 | |
| Deletion | Chr7 | 10390968 | 10390970 | 2 | 2 |
| Deletion | Chr6 | 13332850 | 13332851 | 1 | |
| Deletion | Chr8 | 13583682 | 13583685 | 3 | |
| Deletion | Chr7 | 13632907 | 13632908 | 1 | |
| Deletion | Chr7 | 13789093 | 13789094 | 1 | |
| Deletion | Chr9 | 14215712 | 14215713 | 1 | |
| Deletion | Chr6 | 15428870 | 15429025 | 155 | |
| Deletion | Chr2 | 15696436 | 15696446 | 10 | 2 |
| Deletion | Chr4 | 16259071 | 16259072 | 1 | LOC_Os04g27500 |
| Deletion | Chr6 | 17084368 | 17084419 | 51 | LOC_Os06g29750 |
| Deletion | Chr8 | 18045518 | 18045529 | 11 | |
| Deletion | Chr4 | 18708304 | 18708305 | 1 | |
| Deletion | Chr7 | 19705859 | 19705860 | 1 | |
| Deletion | Chr1 | 20837887 | 20837948 | 61 | |
| Deletion | Chr5 | 21545296 | 21545323 | 27 | |
| Deletion | Chr5 | 23842832 | 23842838 | 6 | |
| Deletion | Chr1 | 24320932 | 24320933 | 1 | 2 |
| Deletion | Chr6 | 24925476 | 24925477 | 1 | |
| Deletion | Chr7 | 26841027 | 26841028 | 1 | |
| Deletion | Chr2 | 27310001 | 27358000 | 47999 | 5 |
| Deletion | Chr2 | 27315001 | 27358000 | 42999 | 5 |
| Deletion | Chr2 | 27625001 | 27645000 | 19999 | 2 |
| Deletion | Chr5 | 27768207 | 27768212 | 5 | |
| Deletion | Chr3 | 28866241 | 28866243 | 2 | |
| Deletion | Chr2 | 28883544 | 28883545 | 1 | |
| Deletion | Chr6 | 29095453 | 29095454 | 1 | |
| Deletion | Chr2 | 30640155 | 30640156 | 1 | |
| Deletion | Chr3 | 35300875 | 35300876 | 1 |
Insertions: 17
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 21322202 | 21322202 | 1 | |
| Insertion | Chr1 | 22425821 | 22425821 | 1 | |
| Insertion | Chr1 | 36294544 | 36294559 | 16 | |
| Insertion | Chr10 | 21822128 | 21822128 | 1 | |
| Insertion | Chr10 | 4422789 | 4422789 | 1 | |
| Insertion | Chr10 | 6889707 | 6889725 | 19 | |
| Insertion | Chr12 | 18133661 | 18133661 | 1 | |
| Insertion | Chr12 | 2792086 | 2792086 | 1 | |
| Insertion | Chr3 | 13027045 | 13027048 | 4 | 2 |
| Insertion | Chr3 | 13027045 | 13027048 | 4 | 2 |
| Insertion | Chr3 | 7180630 | 7180635 | 6 | |
| Insertion | Chr4 | 19115751 | 19115754 | 4 | |
| Insertion | Chr4 | 2631922 | 2631922 | 1 | |
| Insertion | Chr6 | 16041170 | 16041171 | 2 | |
| Insertion | Chr6 | 25377852 | 25377852 | 1 | |
| Insertion | Chr7 | 5041692 | 5041692 | 1 | |
| Insertion | Chr9 | 1383674 | 1383674 | 1 | LOC_Os09g02950 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr12 | 561155 | 629583 | 2 |
| Inversion | Chr12 | 561176 | 749590 | 2 |
Translocations: 23
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 321754 | Chr11 | 305572 | |
| Translocation | Chr12 | 748920 | Chr7 | 13614527 | 2 |
| Translocation | Chr12 | 749587 | Chr7 | 13615241 | 2 |
| Translocation | Chr6 | 1483429 | Chr1 | 3192780 | |
| Translocation | Chr11 | 5131411 | Chr8 | 2543738 | |
| Translocation | Chr12 | 6981603 | Chr3 | 2404419 | LOC_Os12g12664 |
| Translocation | Chr4 | 7859118 | Chr2 | 21394561 | |
| Translocation | Chr6 | 10800746 | Chr4 | 16768921 | |
| Translocation | Chr10 | 11524848 | Chr3 | 19252184 | |
| Translocation | Chr10 | 12201931 | Chr7 | 17150974 | |
| Translocation | Chr9 | 14486469 | Chr6 | 5970921 | |
| Translocation | Chr6 | 15615027 | Chr3 | 25764416 | |
| Translocation | Chr10 | 18195069 | Chr9 | 3016434 | 2 |
| Translocation | Chr12 | 19102087 | Chr5 | 4729804 | |
| Translocation | Chr12 | 19105199 | Chr3 | 14531802 | |
| Translocation | Chr8 | 20160668 | Chr6 | 19700126 | LOC_Os08g32540 |
| Translocation | Chr9 | 20200260 | Chr6 | 14069748 | |
| Translocation | Chr8 | 22388823 | Chr2 | 16788107 | 2 |
| Translocation | Chr6 | 23042479 | Chr3 | 3707180 | LOC_Os03g07270 |
| Translocation | Chr11 | 27850830 | Chr1 | 3225345 | 2 |
| Translocation | Chr11 | 28348469 | Chr1 | 23459456 | |
| Translocation | Chr11 | 28430566 | Chr8 | 5481931 | |
| Translocation | Chr4 | 30957968 | Chr1 | 35222264 |
