Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2719-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2719-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 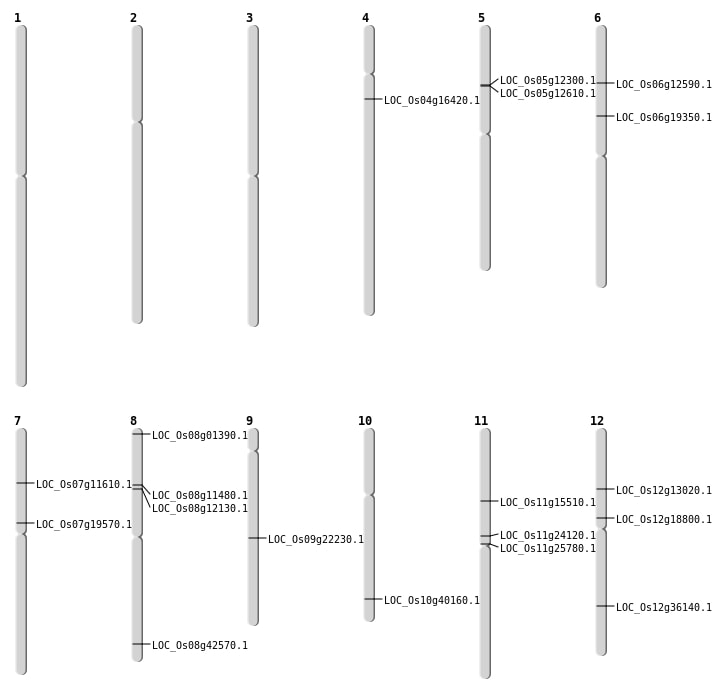
|
| Variant Information |
|---|
Single base substitutions: 85
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 4726833 | C-T | HET | ||
| SBS | Chr1 | 9376583 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr1 | 9853058 | G-A | Homo | ||
| SBS | Chr1 | 9853058 | G-A | HOMO | INTERGENIC | |
| SBS | Chr10 | 13690425 | C-A | HET | ||
| SBS | Chr10 | 14039808 | G-T | HET | ||
| SBS | Chr10 | 19148426 | T-A | HET | ||
| SBS | Chr10 | 19148426 | T-A | HET | UTR_3_PRIME | |
| SBS | Chr10 | 21500310 | C-A | HET | STOP_LOST | LOC_Os10g40160 |
| SBS | Chr10 | 6351727 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 6352729 | T-A | HET | INTERGENIC | |
| SBS | Chr10 | 9962926 | C-A | HET | ||
| SBS | Chr11 | 10770342 | T-C | HET | ||
| SBS | Chr11 | 10770342 | T-C | HET | INTERGENIC | |
| SBS | Chr11 | 14705596 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os11g25780 |
| SBS | Chr11 | 15342906 | A-G | HET | ||
| SBS | Chr11 | 15342906 | A-G | HET | UTR_3_PRIME | |
| SBS | Chr11 | 191158 | C-T | HET | ||
| SBS | Chr11 | 21661551 | A-G | HET | ||
| SBS | Chr11 | 21661551 | A-G | HET | INTERGENIC | |
| SBS | Chr11 | 26487239 | G-A | Homo | ||
| SBS | Chr11 | 26487239 | G-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 26709113 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 8785743 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os11g15510 |
| SBS | Chr12 | 10886791 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os12g18800 |
| SBS | Chr12 | 10886791 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os12g18800 |
| SBS | Chr12 | 12528080 | T-C | HET | INTERGENIC | |
| SBS | Chr12 | 15187143 | G-T | HET | INTERGENIC | |
| SBS | Chr12 | 16401920 | A-C | HET | ||
| SBS | Chr12 | 22073437 | A-T | HET | ||
| SBS | Chr12 | 22153317 | T-G | HET | NON_SYNONYMOUS_CODING | LOC_Os12g36140 |
| SBS | Chr12 | 4584095 | T-A | HET | INTERGENIC | |
| SBS | Chr12 | 7218020 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os12g13020 |
| SBS | Chr12 | 7218020 | G-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os12g13020 |
| SBS | Chr12 | 7675754 | C-T | HET | INTRON | |
| SBS | Chr12 | 7931834 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 10921848 | T-C | HET | ||
| SBS | Chr2 | 10921848 | T-C | HET | INTERGENIC | |
| SBS | Chr2 | 14038746 | C-G | HET | ||
| SBS | Chr2 | 35552139 | T-C | HET | INTRON | |
| SBS | Chr2 | 5668969 | A-G | HET | ||
| SBS | Chr3 | 151486 | G-C | HET | ||
| SBS | Chr3 | 21778063 | T-A | HET | ||
| SBS | Chr3 | 22981035 | G-T | HET | ||
| SBS | Chr3 | 22981035 | G-T | HET | INTERGENIC | |
| SBS | Chr3 | 28243792 | A-G | HET | ||
| SBS | Chr3 | 28243792 | A-G | HET | INTRON | |
| SBS | Chr3 | 30215260 | T-C | Homo | ||
| SBS | Chr3 | 35495343 | T-C | HET | ||
| SBS | Chr4 | 13854963 | A-G | HET | ||
| SBS | Chr4 | 16368897 | A-G | HET | ||
| SBS | Chr4 | 19918787 | T-A | HET | INTERGENIC | |
| SBS | Chr4 | 27366290 | A-G | HET | ||
| SBS | Chr4 | 27706483 | T-A | HET | ||
| SBS | Chr4 | 28627310 | C-T | HET | ||
| SBS | Chr4 | 28627310 | C-T | HOMO | INTRON | |
| SBS | Chr4 | 30937156 | C-A | HET | INTERGENIC | |
| SBS | Chr4 | 8926661 | G-A | HET | STOP_GAINED | LOC_Os04g16420 |
| SBS | Chr4 | 8959179 | C-T | HET | ||
| SBS | Chr4 | 8959179 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 16545878 | A-G | HET | INTRON | |
| SBS | Chr5 | 16925178 | T-A | HET | ||
| SBS | Chr5 | 16925178 | T-A | HET | INTERGENIC | |
| SBS | Chr5 | 17844918 | C-A | HET | ||
| SBS | Chr5 | 17844918 | C-A | HET | INTERGENIC | |
| SBS | Chr5 | 20169241 | C-A | HET | INTERGENIC | |
| SBS | Chr5 | 28116333 | T-C | Homo | ||
| SBS | Chr5 | 28116333 | T-C | HOMO | INTERGENIC | |
| SBS | Chr6 | 11010698 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os06g19350 |
| SBS | Chr6 | 13553381 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 23731550 | C-A | HET | ||
| SBS | Chr7 | 11598160 | T-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os07g19570 |
| SBS | Chr7 | 1288868 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 24531915 | C-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 25481770 | T-A | Homo | ||
| SBS | Chr7 | 25481770 | T-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 6408002 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g11610 |
| SBS | Chr7 | 8660769 | T-C | HET | ||
| SBS | Chr8 | 18170009 | T-C | HET | ||
| SBS | Chr8 | 18170009 | T-C | HET | INTRON | |
| SBS | Chr8 | 19247926 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 19910688 | G-A | HET | ||
| SBS | Chr8 | 253466 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g01390 |
| SBS | Chr9 | 21061546 | C-T | HET | ||
| SBS | Chr9 | 21061546 | C-T | HET | SYNONYMOUS_CODING |
Deletions: 28
Insertions: 21
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 14482109 | 14482122 | 14 | |
| Insertion | Chr1 | 8131394 | 8131395 | 2 | |
| Insertion | Chr10 | 22711146 | 22711147 | 2 | 2 |
| Insertion | Chr10 | 22711146 | 22711148 | 3 | 2 |
| Insertion | Chr10 | 7145753 | 7145753 | 1 | |
| Insertion | Chr12 | 18990627 | 18990628 | 2 | |
| Insertion | Chr12 | 2939560 | 2939575 | 16 | |
| Insertion | Chr2 | 11590335 | 11590338 | 4 | |
| Insertion | Chr2 | 30639797 | 30639797 | 1 | |
| Insertion | Chr3 | 14316094 | 14316095 | 2 | |
| Insertion | Chr3 | 28037278 | 28037279 | 2 | |
| Insertion | Chr4 | 28015059 | 28015060 | 2 | 2 |
| Insertion | Chr4 | 28015059 | 28015060 | 2 | 2 |
| Insertion | Chr4 | 7947891 | 7947891 | 1 | |
| Insertion | Chr5 | 29931151 | 29931151 | 1 | |
| Insertion | Chr6 | 9464981 | 9464987 | 7 | 2 |
| Insertion | Chr6 | 9464981 | 9464987 | 7 | 2 |
| Insertion | Chr8 | 11049434 | 11049434 | 1 | |
| Insertion | Chr8 | 16867262 | 16867262 | 1 | |
| Insertion | Chr8 | 19211524 | 19211525 | 2 | |
| Insertion | Chr9 | 14315063 | 14315066 | 4 |
Inversions: 5
Translocations: 14
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr10 | 2265231 | Chr9 | 5503275 | |
| Translocation | Chr3 | 6780070 | Chr2 | 12010625 | |
| Translocation | Chr6 | 6823006 | Chr4 | 27350794 | LOC_Os06g12590 |
| Translocation | Chr11 | 7720116 | Chr6 | 4358327 | |
| Translocation | Chr2 | 8310769 | Chr1 | 31296068 | |
| Translocation | Chr10 | 8620598 | Chr1 | 12472002 | |
| Translocation | Chr9 | 11816226 | Chr2 | 6533224 | |
| Translocation | Chr12 | 12883131 | Chr10 | 4481271 | |
| Translocation | Chr9 | 13448269 | Chr8 | 7145853 | 2 |
| Translocation | Chr11 | 13708396 | Chr6 | 11097876 | LOC_Os11g24120 |
| Translocation | Chr9 | 15005716 | Chr8 | 1594438 | |
| Translocation | Chr9 | 16564127 | Chr1 | 24311693 | |
| Translocation | Chr11 | 19693914 | Chr2 | 5291366 | |
| Translocation | Chr8 | 26906514 | Chr3 | 25348841 | LOC_Os08g42570 |
