Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2723-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2723-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 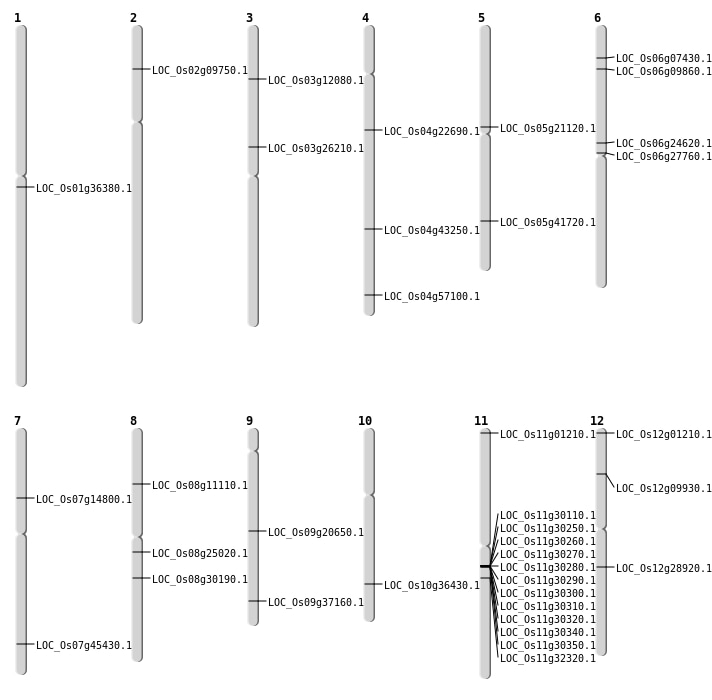
|
| Variant Information |
|---|
Single base substitutions: 92
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 16851299 | C-T | HET | ||
| SBS | Chr1 | 17077485 | C-A | HET | INTERGENIC | |
| SBS | Chr1 | 18255199 | G-A | HET | INTRON | |
| SBS | Chr1 | 20247758 | G-A | HET | ||
| SBS | Chr1 | 20247758 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 30841359 | C-T | HET | ||
| SBS | Chr1 | 30841359 | C-T | HET | INTRON | |
| SBS | Chr1 | 32936458 | A-T | HET | INTERGENIC | |
| SBS | Chr1 | 34012468 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 40279793 | T-A | HET | INTERGENIC | |
| SBS | Chr10 | 18791917 | A-T | HET | ||
| SBS | Chr10 | 18791917 | A-T | HET | INTERGENIC | |
| SBS | Chr10 | 18843404 | T-A | HET | ||
| SBS | Chr10 | 22149372 | C-T | HET | ||
| SBS | Chr10 | 325354 | G-A | HET | ||
| SBS | Chr10 | 4512937 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 10175029 | C-T | HET | ||
| SBS | Chr11 | 10175029 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 12761990 | C-A | HET | ||
| SBS | Chr11 | 22081896 | C-A | HET | ||
| SBS | Chr11 | 9943259 | G-T | HET | ||
| SBS | Chr12 | 1023814 | A-T | Homo | ||
| SBS | Chr12 | 1023814 | A-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 13606442 | A-C | HET | INTERGENIC | |
| SBS | Chr12 | 13924539 | T-C | HET | ||
| SBS | Chr12 | 14508232 | C-T | HET | ||
| SBS | Chr12 | 14508232 | C-T | HET | INTRON | |
| SBS | Chr12 | 15613360 | A-G | Homo | ||
| SBS | Chr12 | 15613360 | A-G | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr12 | 16327320 | A-G | HET | ||
| SBS | Chr12 | 16518102 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 3054935 | C-A | HET | ||
| SBS | Chr12 | 3054935 | C-A | HET | INTERGENIC | |
| SBS | Chr2 | 11876071 | C-T | HET | ||
| SBS | Chr2 | 139166 | T-A | HET | INTERGENIC | |
| SBS | Chr2 | 139168 | C-G | HET | INTERGENIC | |
| SBS | Chr2 | 15419543 | G-T | HET | INTERGENIC | |
| SBS | Chr2 | 16312423 | C-T | HET | ||
| SBS | Chr2 | 16312423 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 21751281 | G-A | HET | ||
| SBS | Chr2 | 21751281 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 23021799 | G-A | HET | ||
| SBS | Chr2 | 2424846 | C-G | HET | ||
| SBS | Chr2 | 2424846 | C-G | HET | INTERGENIC | |
| SBS | Chr2 | 28971528 | C-A | HET | INTRON | |
| SBS | Chr2 | 4287145 | G-A | Homo | ||
| SBS | Chr2 | 4287145 | G-A | HOMO | UTR_3_PRIME | |
| SBS | Chr3 | 1519129 | G-A | Homo | ||
| SBS | Chr3 | 1519129 | G-A | HOMO | UTR_5_PRIME | |
| SBS | Chr3 | 21988069 | G-T | Homo | ||
| SBS | Chr3 | 21988069 | G-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 28809447 | T-G | HET | ||
| SBS | Chr3 | 34421284 | G-T | HET | INTERGENIC | |
| SBS | Chr3 | 6340620 | G-A | HET | STOP_GAINED | LOC_Os03g12080 |
| SBS | Chr4 | 12521245 | G-A | Homo | ||
| SBS | Chr4 | 12521245 | G-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 12521246 | G-A | Homo | ||
| SBS | Chr4 | 12521246 | G-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 12859413 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g22690 |
| SBS | Chr4 | 20382700 | C-T | HET | INTRON | |
| SBS | Chr4 | 2510417 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 25590116 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g43250 |
| SBS | Chr4 | 34025892 | G-A | HET | STOP_GAINED | LOC_Os04g57100 |
| SBS | Chr5 | 12230112 | C-A | HET | INTERGENIC | |
| SBS | Chr5 | 18184354 | G-A | HET | ||
| SBS | Chr5 | 18184354 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 22889438 | C-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 24410027 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g41720 |
| SBS | Chr5 | 2750587 | C-A | HET | ||
| SBS | Chr5 | 29486665 | T-C | HET | ||
| SBS | Chr6 | 13101046 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 19682220 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 14690442 | C-T | HET | ||
| SBS | Chr7 | 14690442 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 17093962 | A-G | HET | ||
| SBS | Chr7 | 17093962 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 21679367 | T-G | HET | ||
| SBS | Chr7 | 21679367 | T-G | HET | INTERGENIC | |
| SBS | Chr7 | 26081899 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 454764 | G-A | Homo | ||
| SBS | Chr7 | 454764 | G-A | HOMO | INTRON | |
| SBS | Chr8 | 12449521 | T-C | HET | ||
| SBS | Chr8 | 18559385 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g30190 |
| SBS | Chr8 | 5411434 | G-A | HET | ||
| SBS | Chr8 | 6258402 | G-A | HET | ||
| SBS | Chr8 | 7424333 | G-A | HET | INTRON | |
| SBS | Chr9 | 14986632 | A-T | HET | ||
| SBS | Chr9 | 14986632 | A-T | HET | INTERGENIC | |
| SBS | Chr9 | 17876182 | A-C | HET | ||
| SBS | Chr9 | 17876182 | A-C | HET | INTRON | |
| SBS | Chr9 | 2902222 | G-T | HET | ||
| SBS | Chr9 | 8179897 | C-G | HET |
Deletions: 30
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr6 | 3571986 | 3571987 | 1 | 2 |
| Deletion | Chr12 | 5259815 | 5259816 | 1 | LOC_Os12g09930 |
| Deletion | Chr4 | 6059245 | 6059252 | 7 | 2 |
| Deletion | Chr4 | 6670807 | 6670808 | 1 | |
| Deletion | Chr2 | 7832011 | 7832129 | 118 | |
| Deletion | Chr7 | 8452729 | 8452730 | 1 | LOC_Os07g14800 |
| Deletion | Chr2 | 9005438 | 9005440 | 2 | |
| Deletion | Chr11 | 11722420 | 11722421 | 1 | |
| Deletion | Chr3 | 13348650 | 13348702 | 52 | |
| Deletion | Chr3 | 15001483 | 15001485 | 2 | LOC_Os03g26210 |
| Deletion | Chr4 | 15002607 | 15002608 | 1 | |
| Deletion | Chr8 | 15189784 | 15189785 | 1 | LOC_Os08g25020 |
| Deletion | Chr12 | 15373681 | 15373682 | 1 | |
| Deletion | Chr12 | 16898154 | 16898156 | 2 | |
| Deletion | Chr11 | 17583001 | 17619000 | 35999 | 10 |
| Deletion | Chr11 | 17585001 | 17618000 | 32999 | 10 |
| Deletion | Chr12 | 18801751 | 18801756 | 5 | |
| Deletion | Chr10 | 19320924 | 19320931 | 7 | |
| Deletion | Chr10 | 19494605 | 19494606 | 1 | LOC_Os10g36430 |
| Deletion | Chr1 | 20188117 | 20188206 | 89 | LOC_Os01g36380 |
| Deletion | Chr3 | 20391081 | 20391082 | 1 | |
| Deletion | Chr12 | 20827807 | 20827829 | 22 | 2 |
| Deletion | Chr9 | 21435383 | 21435399 | 16 | LOC_Os09g37160 |
| Deletion | Chr1 | 22241353 | 22241359 | 6 | |
| Deletion | Chr4 | 26161210 | 26161218 | 8 | |
| Deletion | Chr5 | 26187546 | 26187558 | 12 | |
| Deletion | Chr6 | 28417945 | 28417949 | 4 | |
| Deletion | Chr4 | 29806195 | 29806201 | 6 | |
| Deletion | Chr1 | 33576556 | 33576557 | 1 | |
| Deletion | Chr1 | 42997097 | 42997104 | 7 |
Insertions: 19
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 31933798 | 31933798 | 1 | |
| Insertion | Chr11 | 20993894 | 20993895 | 2 | |
| Insertion | Chr12 | 17096932 | 17096932 | 1 | LOC_Os12g28920 |
| Insertion | Chr2 | 25476436 | 25476436 | 1 | |
| Insertion | Chr2 | 6710927 | 6710934 | 8 | |
| Insertion | Chr3 | 19759561 | 19759561 | 1 | |
| Insertion | Chr3 | 2292369 | 2292370 | 2 | |
| Insertion | Chr4 | 10182469 | 10182469 | 1 | |
| Insertion | Chr6 | 1026777 | 1026778 | 2 | 2 |
| Insertion | Chr6 | 1026777 | 1026778 | 2 | 2 |
| Insertion | Chr6 | 12551051 | 12551051 | 1 | |
| Insertion | Chr6 | 15256408 | 15256423 | 16 | |
| Insertion | Chr6 | 27094509 | 27094510 | 2 | 2 |
| Insertion | Chr6 | 27094509 | 27094510 | 2 | 2 |
| Insertion | Chr8 | 13844239 | 13844239 | 1 | |
| Insertion | Chr8 | 24362510 | 24362510 | 1 | |
| Insertion | Chr8 | 26736967 | 26736976 | 10 | |
| Insertion | Chr8 | 6448074 | 6448074 | 1 | |
| Insertion | Chr8 | 9568995 | 9568995 | 1 |
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr2 | 4700111 | 5029761 | 2 |
| Inversion | Chr2 | 4700121 | 5029775 | 2 |
| Inversion | Chr6 | 5018083 | 5018183 | LOC_Os06g09860 |
| Inversion | Chr12 | 11921444 | 11922062 |
Translocations: 20
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 127738 | Chr11 | 128897 | 2 |
| Translocation | Chr9 | 2592229 | Chr7 | 27101163 | 2 |
| Translocation | Chr9 | 4920900 | Chr1 | 19588631 | |
| Translocation | Chr10 | 5298646 | Chr8 | 14523301 | |
| Translocation | Chr11 | 5743800 | Chr10 | 12207983 | |
| Translocation | Chr8 | 6538918 | Chr4 | 3094411 | LOC_Os08g11110 |
| Translocation | Chr10 | 8097551 | Chr1 | 16931745 | |
| Translocation | Chr7 | 9387189 | Chr4 | 20072514 | |
| Translocation | Chr6 | 10117993 | Chr5 | 21218472 | |
| Translocation | Chr9 | 12438833 | Chr3 | 17343867 | 2 |
| Translocation | Chr9 | 12438834 | Chr3 | 17344354 | 2 |
| Translocation | Chr8 | 13038720 | Chr7 | 14407536 | |
| Translocation | Chr6 | 14457838 | Chr4 | 8094447 | LOC_Os06g24620 |
| Translocation | Chr11 | 15613259 | Chr9 | 749691 | |
| Translocation | Chr3 | 16812701 | Chr1 | 18860178 | |
| Translocation | Chr11 | 17509681 | Chr2 | 944616 | LOC_Os11g30110 |
| Translocation | Chr11 | 17510217 | Chr2 | 944616 | |
| Translocation | Chr12 | 24881287 | Chr6 | 15720880 | 2 |
| Translocation | Chr12 | 24881292 | Chr5 | 12422855 | 2 |
| Translocation | Chr12 | 27114959 | Chr3 | 17206796 |
