Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2724-S-1 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2724-S-1 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 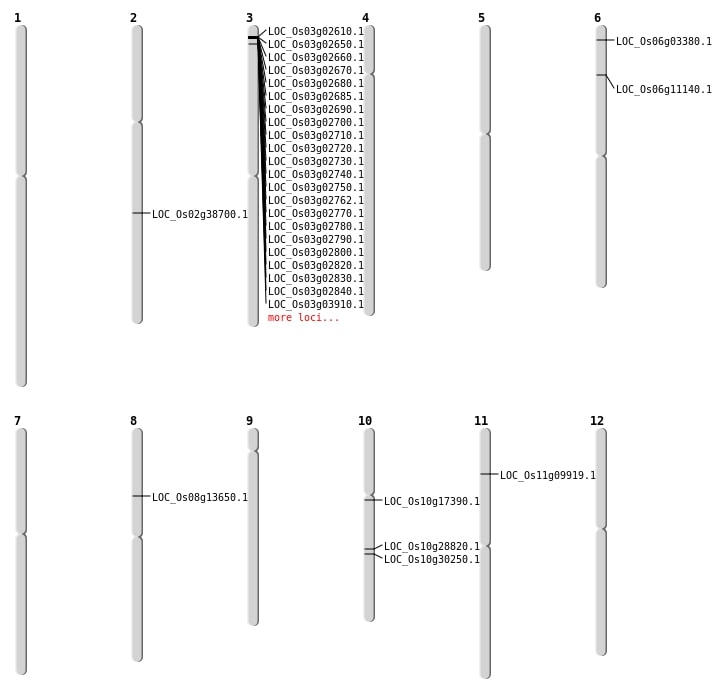
|
| Variant Information |
|---|
Single base substitutions: 35
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 12823354 | A-G | HET | INTERGENIC | |
| SBS | Chr1 | 16716394 | T-A | HET | INTERGENIC | |
| SBS | Chr1 | 24900792 | C-G | HET | SYNONYMOUS_CODING | |
| SBS | Chr1 | 42289672 | A-G | HET | INTERGENIC | |
| SBS | Chr1 | 42645991 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 15026475 | T-C | HOMO | NON_SYNONYMOUS_CODING | LOC_Os10g28820 |
| SBS | Chr10 | 9183788 | A-G | HOMO | INTERGENIC | |
| SBS | Chr11 | 15025330 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 14972853 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 29130293 | C-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 29201436 | T-A | HOMO | INTRON | |
| SBS | Chr2 | 29201437 | G-A | HOMO | INTRON | |
| SBS | Chr3 | 19325189 | C-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr3 | 20998327 | T-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 27502209 | T-C | HET | SPLICE_SITE_REGION | |
| SBS | Chr3 | 28285990 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 29497643 | C-A | HET | INTERGENIC | |
| SBS | Chr3 | 30934365 | A-C | HET | INTERGENIC | |
| SBS | Chr4 | 13545714 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr4 | 1369314 | A-G | HET | INTERGENIC | |
| SBS | Chr4 | 22563852 | A-G | HET | INTRON | |
| SBS | Chr5 | 19887232 | C-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 19887233 | A-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 11285826 | T-A | HET | INTRON | |
| SBS | Chr6 | 1290743 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os06g03380 |
| SBS | Chr6 | 1872307 | C-A | HET | INTERGENIC | |
| SBS | Chr6 | 1872308 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 29066343 | T-A | HET | INTERGENIC | |
| SBS | Chr7 | 12567583 | T-C | HET | INTRON | |
| SBS | Chr8 | 18273947 | G-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 20680692 | A-T | HOMO | INTERGENIC | |
| SBS | Chr8 | 2333791 | A-G | HET | INTERGENIC | |
| SBS | Chr8 | 2542960 | G-C | HET | INTERGENIC | |
| SBS | Chr8 | 8120347 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os08g13650 |
| SBS | Chr9 | 1342413 | A-T | HET | INTERGENIC |
Deletions: 26
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr7 | 314092 | 314094 | 2 | |
| Deletion | Chr10 | 701702 | 701703 | 1 | |
| Deletion | Chr3 | 971001 | 1118000 | 146999 | 21 |
| Deletion | Chr6 | 1114001 | 1123000 | 8999 | LOC_Os06g11140 |
| Deletion | Chr1 | 2607533 | 2607537 | 4 | |
| Deletion | Chr4 | 2797684 | 2797685 | 1 | |
| Deletion | Chr10 | 4101050 | 4101052 | 2 | |
| Deletion | Chr9 | 5768749 | 5768767 | 18 | |
| Deletion | Chr8 | 6683197 | 6683198 | 1 | |
| Deletion | Chr7 | 7250670 | 7250691 | 21 | |
| Deletion | Chr4 | 7293763 | 7293764 | 1 | |
| Deletion | Chr10 | 8763658 | 8763674 | 16 | LOC_Os10g17390 |
| Deletion | Chr2 | 9198111 | 9198112 | 1 | |
| Deletion | Chr12 | 10311523 | 10311525 | 2 | |
| Deletion | Chr7 | 15090996 | 15090997 | 1 | |
| Deletion | Chr5 | 15279212 | 15279213 | 1 | |
| Deletion | Chr11 | 15615272 | 15615275 | 3 | |
| Deletion | Chr10 | 15714837 | 15714839 | 2 | LOC_Os10g30250 |
| Deletion | Chr1 | 18634078 | 18634113 | 35 | |
| Deletion | Chr12 | 19021652 | 19021665 | 13 | |
| Deletion | Chr7 | 19989878 | 19989890 | 12 | |
| Deletion | Chr11 | 26727910 | 26727911 | 1 | |
| Deletion | Chr6 | 29273253 | 29273254 | 1 | |
| Deletion | Chr4 | 30464968 | 30464971 | 3 | |
| Deletion | Chr3 | 31357705 | 31357709 | 4 | |
| Deletion | Chr1 | 33352458 | 33352461 | 3 |
Insertions: 12
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 1307789 | 1307789 | 1 | |
| Insertion | Chr1 | 27072046 | 27072046 | 1 | |
| Insertion | Chr12 | 15324301 | 15324332 | 32 | |
| Insertion | Chr12 | 22275126 | 22275126 | 1 | |
| Insertion | Chr2 | 23393977 | 23393978 | 2 | LOC_Os02g38700 |
| Insertion | Chr3 | 970242 | 970242 | 1 | |
| Insertion | Chr4 | 12174090 | 12174126 | 37 | |
| Insertion | Chr4 | 2952709 | 2952709 | 1 | |
| Insertion | Chr6 | 16047192 | 16047260 | 69 | |
| Insertion | Chr6 | 25226022 | 25226115 | 94 | |
| Insertion | Chr7 | 20991174 | 20991177 | 4 | |
| Insertion | Chr8 | 13869200 | 13869200 | 1 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr3 | 964968 | 1789128 | 2 |
| Inversion | Chr3 | 1118444 | 1507903 |
Translocations: 4
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr7 | 4765762 | Chr1 | 40659048 | |
| Translocation | Chr11 | 5309499 | Chr10 | 16682476 | LOC_Os11g09919 |
| Translocation | Chr11 | 5309736 | Chr10 | 16682459 | LOC_Os11g09919 |
| Translocation | Chr11 | 7720116 | Chr6 | 4358308 |
