Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2725-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2725-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 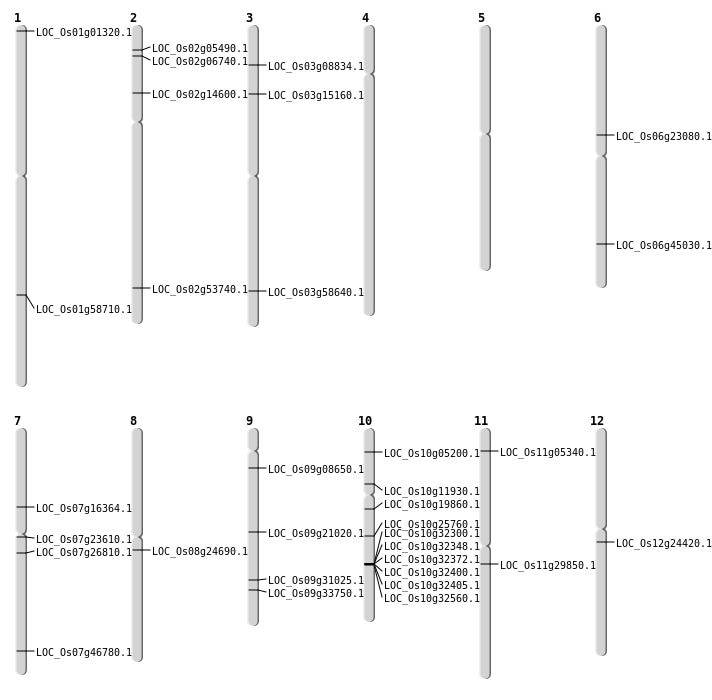
|
| Variant Information |
|---|
Single base substitutions: 132
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 11756568 | C-T | HET | ||
| SBS | Chr1 | 11756568 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 1251589 | A-G | Homo | ||
| SBS | Chr1 | 15572817 | C-T | Homo | ||
| SBS | Chr1 | 15572817 | C-T | HOMO | INTRON | |
| SBS | Chr1 | 17010469 | A-G | Homo | ||
| SBS | Chr1 | 17010469 | A-G | HOMO | INTERGENIC | |
| SBS | Chr1 | 17135189 | T-G | Homo | ||
| SBS | Chr1 | 17135189 | T-G | HOMO | INTERGENIC | |
| SBS | Chr1 | 33073022 | C-T | HET | ||
| SBS | Chr1 | 33073022 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 33162737 | C-G | HET | ||
| SBS | Chr1 | 33162737 | C-G | HET | INTERGENIC | |
| SBS | Chr1 | 33942994 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g58710 |
| SBS | Chr1 | 34392738 | C-T | HET | ||
| SBS | Chr1 | 34392738 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 40884064 | C-T | HET | ||
| SBS | Chr1 | 40884064 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 4177322 | C-A | HET | ||
| SBS | Chr10 | 10179376 | A-G | Homo | ||
| SBS | Chr10 | 10179376 | A-G | HOMO | INTERGENIC | |
| SBS | Chr10 | 12051403 | T-A | Homo | ||
| SBS | Chr10 | 12051403 | T-A | HOMO | INTRON | |
| SBS | Chr10 | 16967570 | T-C | HOMO | INTERGENIC | |
| SBS | Chr10 | 20563734 | C-A | Homo | ||
| SBS | Chr10 | 20563734 | C-A | HOMO | INTERGENIC | |
| SBS | Chr10 | 22054887 | G-T | Homo | ||
| SBS | Chr10 | 22054887 | G-T | HOMO | INTERGENIC | |
| SBS | Chr10 | 3322159 | C-T | HET | ||
| SBS | Chr10 | 3328261 | G-C | Homo | ||
| SBS | Chr10 | 3328261 | G-C | HOMO | INTERGENIC | |
| SBS | Chr10 | 6707478 | A-G | Homo | ||
| SBS | Chr10 | 6707478 | A-G | HOMO | INTERGENIC | |
| SBS | Chr10 | 9049350 | C-T | Homo | ||
| SBS | Chr10 | 9049350 | C-T | HOMO | INTRON | |
| SBS | Chr11 | 14531401 | G-T | HET | ||
| SBS | Chr11 | 14531401 | G-T | HET | INTERGENIC | |
| SBS | Chr11 | 17331039 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os11g29850 |
| SBS | Chr11 | 23639822 | C-T | HET | ||
| SBS | Chr11 | 23639822 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 2366329 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g05340 |
| SBS | Chr12 | 12262962 | G-C | HET | ||
| SBS | Chr12 | 12262962 | G-C | HET | INTERGENIC | |
| SBS | Chr12 | 18061438 | T-C | HET | INTERGENIC | |
| SBS | Chr12 | 22115824 | C-T | HET | ||
| SBS | Chr12 | 22115824 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 3472955 | C-T | HET | ||
| SBS | Chr12 | 6225119 | G-A | HET | ||
| SBS | Chr12 | 6225119 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 15920171 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 23658613 | A-T | Homo | ||
| SBS | Chr2 | 23658613 | A-T | HOMO | INTRON | |
| SBS | Chr2 | 25670941 | A-C | Homo | ||
| SBS | Chr2 | 25670941 | A-C | HOMO | INTRON | |
| SBS | Chr2 | 33274863 | C-T | HET | ||
| SBS | Chr2 | 33274863 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 7171336 | A-G | Homo | ||
| SBS | Chr2 | 7171336 | A-G | HOMO | INTERGENIC | |
| SBS | Chr2 | 7502431 | G-C | Homo | ||
| SBS | Chr2 | 7502431 | G-C | HOMO | INTRON | |
| SBS | Chr3 | 15080629 | C-T | HET | ||
| SBS | Chr3 | 15080629 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr3 | 20556672 | A-T | HET | ||
| SBS | Chr3 | 20556672 | A-T | HET | INTRON | |
| SBS | Chr3 | 2537993 | G-A | HET | ||
| SBS | Chr3 | 2537993 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 26893589 | C-A | HET | INTRON | |
| SBS | Chr3 | 27409767 | A-C | HET | ||
| SBS | Chr3 | 27409767 | A-C | HET | INTRON | |
| SBS | Chr3 | 30974030 | T-C | HET | ||
| SBS | Chr3 | 30974030 | T-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr3 | 33399366 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os03g58640 |
| SBS | Chr3 | 35587572 | G-A | HET | ||
| SBS | Chr3 | 8710033 | C-T | Homo | ||
| SBS | Chr3 | 8710033 | C-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 13106144 | A-T | HET | INTERGENIC | |
| SBS | Chr4 | 22576125 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 23800692 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 24486051 | G-T | HET | ||
| SBS | Chr4 | 24486051 | G-T | HET | INTRON | |
| SBS | Chr4 | 30138716 | C-A | HET | ||
| SBS | Chr4 | 30138716 | C-A | HET | INTERGENIC | |
| SBS | Chr4 | 34736713 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 4896678 | A-T | HET | ||
| SBS | Chr4 | 8985558 | A-C | HET | ||
| SBS | Chr4 | 8985558 | A-C | HET | INTRON | |
| SBS | Chr5 | 14260461 | G-A | HET | ||
| SBS | Chr5 | 14260461 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 18225851 | C-T | Homo | ||
| SBS | Chr5 | 18225851 | C-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 19150799 | T-C | HET | ||
| SBS | Chr5 | 28348596 | T-C | HET | INTERGENIC | |
| SBS | Chr5 | 5064534 | C-G | HET | ||
| SBS | Chr6 | 15568844 | A-T | HET | ||
| SBS | Chr6 | 15568844 | A-T | HET | INTRON | |
| SBS | Chr6 | 15568845 | T-A | HET | ||
| SBS | Chr6 | 15568845 | T-A | HET | INTRON | |
| SBS | Chr6 | 17740750 | C-T | HET | ||
| SBS | Chr6 | 20088079 | C-A | HET | ||
| SBS | Chr6 | 20088079 | C-A | HET | UTR_3_PRIME | |
| SBS | Chr6 | 4002638 | G-A | HET | ||
| SBS | Chr6 | 448797 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 9936920 | G-T | HET | ||
| SBS | Chr7 | 18754915 | G-A | Homo | ||
| SBS | Chr7 | 18754915 | G-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 23496658 | C-T | HET | ||
| SBS | Chr7 | 23571070 | G-A | Homo | ||
| SBS | Chr7 | 23571070 | G-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 26750819 | A-G | Homo | ||
| SBS | Chr7 | 26750819 | A-G | HOMO | INTERGENIC | |
| SBS | Chr7 | 27956711 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g46780 |
| SBS | Chr7 | 7162950 | T-C | HET | ||
| SBS | Chr7 | 7162950 | T-C | HET | INTERGENIC | |
| SBS | Chr7 | 8544040 | C-T | HET | SYNONYMOUS_STOP | |
| SBS | Chr8 | 17128570 | T-C | HET | ||
| SBS | Chr8 | 17128570 | T-C | HET | INTRON | |
| SBS | Chr8 | 20892264 | C-T | HET | ||
| SBS | Chr8 | 23436547 | A-T | Homo | ||
| SBS | Chr8 | 23436547 | A-T | HOMO | INTERGENIC | |
| SBS | Chr8 | 8503200 | T-C | HET | ||
| SBS | Chr8 | 8503200 | T-C | HET | INTERGENIC | |
| SBS | Chr9 | 15965745 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 17483253 | T-C | Homo | ||
| SBS | Chr9 | 17483253 | T-C | HOMO | INTERGENIC | |
| SBS | Chr9 | 1926523 | G-A | HET | ||
| SBS | Chr9 | 1969887 | C-T | HET | ||
| SBS | Chr9 | 1969887 | C-T | HET | INTRON | |
| SBS | Chr9 | 22374907 | T-C | HET | ||
| SBS | Chr9 | 22374907 | T-C | HET | INTERGENIC | |
| SBS | Chr9 | 2715111 | G-T | HET | INTERGENIC | |
| SBS | Chr9 | 4530675 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os09g08650 |
| SBS | Chr9 | 5008652 | C-A | HET |
Deletions: 41
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr2 | 368842 | 368843 | 1 | |
| Deletion | Chr7 | 972891 | 972892 | 1 | |
| Deletion | Chr12 | 1590810 | 1590816 | 6 | 2 |
| Deletion | Chr5 | 2274830 | 2274831 | 1 | |
| Deletion | Chr10 | 2549515 | 2549519 | 4 | 2 |
| Deletion | Chr2 | 2656475 | 2656478 | 3 | 2 |
| Deletion | Chr10 | 3336041 | 3336054 | 13 | 2 |
| Deletion | Chr2 | 3394477 | 3394482 | 5 | 2 |
| Deletion | Chr3 | 4172910 | 4172911 | 1 | 2 |
| Deletion | Chr4 | 4849085 | 4849156 | 71 | |
| Deletion | Chr7 | 5301251 | 5301270 | 19 | |
| Deletion | Chr8 | 6341654 | 6341733 | 79 | |
| Deletion | Chr9 | 7335141 | 7335239 | 98 | |
| Deletion | Chr2 | 8058748 | 8058749 | 1 | 2 |
| Deletion | Chr2 | 8177575 | 8177576 | 1 | 2 |
| Deletion | Chr3 | 8285370 | 8285371 | 1 | LOC_Os03g15160 |
| Deletion | Chr1 | 8481130 | 8481145 | 15 | 2 |
| Deletion | Chr6 | 9711298 | 9711302 | 4 | |
| Deletion | Chr5 | 10198056 | 10198057 | 1 | |
| Deletion | Chr3 | 10393192 | 10393194 | 2 | 2 |
| Deletion | Chr6 | 11800947 | 11800952 | 5 | |
| Deletion | Chr9 | 12679376 | 12679377 | 1 | LOC_Os09g21020 |
| Deletion | Chr10 | 13347357 | 13347359 | 2 | 2 |
| Deletion | Chr7 | 13347508 | 13347509 | 1 | LOC_Os07g23610 |
| Deletion | Chr7 | 13690648 | 13690653 | 5 | |
| Deletion | Chr8 | 14916032 | 14916038 | 6 | 2 |
| Deletion | Chr7 | 15493504 | 15493516 | 12 | 2 |
| Deletion | Chr7 | 16137225 | 16137237 | 12 | |
| Deletion | Chr10 | 16950001 | 16991000 | 40999 | 5 |
| Deletion | Chr10 | 16952001 | 16991000 | 38999 | 5 |
| Deletion | Chr5 | 17263579 | 17263592 | 13 | 2 |
| Deletion | Chr7 | 17405943 | 17405945 | 2 | 2 |
| Deletion | Chr7 | 18694457 | 18694467 | 10 | |
| Deletion | Chr9 | 19936471 | 19936474 | 3 | LOC_Os09g33750 |
| Deletion | Chr11 | 20235107 | 20235108 | 1 | |
| Deletion | Chr2 | 25503357 | 25503369 | 12 | 2 |
| Deletion | Chr6 | 27244662 | 27244664 | 2 | 2 |
| Deletion | Chr3 | 27328619 | 27328620 | 1 | |
| Deletion | Chr11 | 28405922 | 28406014 | 92 | |
| Deletion | Chr4 | 30859493 | 30859495 | 2 | |
| Deletion | Chr1 | 40770942 | 40770943 | 1 |
Insertions: 19
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 158016 | 158016 | 1 | LOC_Os01g01320 |
| Insertion | Chr1 | 7065368 | 7065373 | 6 | |
| Insertion | Chr1 | 9866617 | 9866618 | 2 | 2 |
| Insertion | Chr1 | 9866617 | 9866618 | 2 | 2 |
| Insertion | Chr10 | 5883741 | 5883741 | 1 | |
| Insertion | Chr11 | 13254646 | 13254646 | 1 | |
| Insertion | Chr11 | 14145230 | 14145230 | 1 | |
| Insertion | Chr11 | 23155758 | 23155763 | 6 | |
| Insertion | Chr11 | 9987199 | 9987199 | 1 | |
| Insertion | Chr12 | 24906263 | 24906263 | 1 | |
| Insertion | Chr2 | 30175401 | 30175403 | 3 | |
| Insertion | Chr2 | 34434078 | 34434078 | 1 | |
| Insertion | Chr3 | 9570690 | 9570697 | 8 | |
| Insertion | Chr4 | 10182469 | 10182469 | 1 | |
| Insertion | Chr4 | 7706796 | 7706796 | 1 | |
| Insertion | Chr6 | 11098499 | 11098499 | 1 | |
| Insertion | Chr8 | 1697012 | 1697015 | 4 | |
| Insertion | Chr8 | 25824153 | 25824154 | 2 | 2 |
| Insertion | Chr8 | 25824153 | 25824154 | 2 | 2 |
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr1 | 2981348 | 2981541 |
Translocations: 14
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr10 | 2470738 | Chr4 | 679517 | |
| Translocation | Chr10 | 6618984 | Chr6 | 13457404 | 2 |
| Translocation | Chr10 | 6928220 | Chr8 | 19394181 | |
| Translocation | Chr10 | 9912215 | Chr2 | 32908783 | 2 |
| Translocation | Chr11 | 9960409 | Chr8 | 22924000 | |
| Translocation | Chr7 | 10706080 | Chr3 | 4570858 | LOC_Os03g08834 |
| Translocation | Chr11 | 12631453 | Chr10 | 4408877 | |
| Translocation | Chr10 | 13030905 | Chr9 | 18669036 | |
| Translocation | Chr10 | 13030909 | Chr9 | 18668209 | 2 |
| Translocation | Chr12 | 13936898 | Chr4 | 16502356 | LOC_Os12g24420 |
| Translocation | Chr8 | 19194685 | Chr7 | 9565473 | 2 |
| Translocation | Chr8 | 19691030 | Chr7 | 9266819 | |
| Translocation | Chr7 | 20402996 | Chr6 | 29925132 | |
| Translocation | Chr7 | 25340100 | Chr3 | 31170074 |
