Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2726-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2726-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 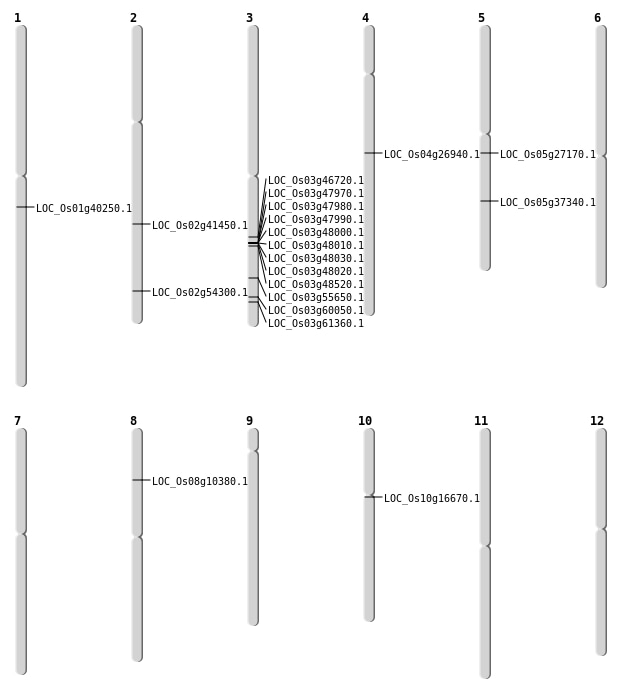
|
| Variant Information |
|---|
Single base substitutions: 45
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 17765234 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 21375444 | A-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr1 | 21378812 | C-T | HOMO | INTRON | |
| SBS | Chr1 | 22725927 | T-C | HOMO | NON_SYNONYMOUS_CODING | LOC_Os01g40250 |
| SBS | Chr1 | 28427909 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 35236202 | G-T | HET | INTERGENIC | |
| SBS | Chr1 | 36525248 | A-G | HET | INTERGENIC | |
| SBS | Chr1 | 41577467 | G-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr1 | 41726711 | T-A | HOMO | INTERGENIC | |
| SBS | Chr10 | 10985501 | T-A | HET | INTERGENIC | |
| SBS | Chr10 | 11420372 | A-T | HET | SPLICE_SITE_REGION | |
| SBS | Chr10 | 12206886 | T-C | HET | INTERGENIC | |
| SBS | Chr11 | 9200123 | A-G | HOMO | INTERGENIC | |
| SBS | Chr12 | 14369242 | C-T | HET | INTRON | |
| SBS | Chr12 | 27207326 | C-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 10634336 | C-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 13024414 | C-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 15903534 | C-T | HOMO | INTRON | |
| SBS | Chr2 | 33300096 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os02g54300 |
| SBS | Chr2 | 5379539 | G-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr3 | 2501667 | A-T | HOMO | INTRON | |
| SBS | Chr3 | 26440700 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os03g46720 |
| SBS | Chr3 | 29378168 | A-C | HET | INTERGENIC | |
| SBS | Chr3 | 2953758 | A-C | HOMO | INTERGENIC | |
| SBS | Chr3 | 30118070 | T-A | HET | INTERGENIC | |
| SBS | Chr3 | 31688651 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os03g55650 |
| SBS | Chr3 | 34826020 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os03g61360 |
| SBS | Chr3 | 34826022 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g61360 |
| SBS | Chr4 | 13682291 | A-T | HET | INTERGENIC | |
| SBS | Chr4 | 15937864 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g26940 |
| SBS | Chr4 | 21827385 | A-G | HET | SYNONYMOUS_CODING | |
| SBS | Chr4 | 22751005 | A-G | HET | INTRON | |
| SBS | Chr5 | 15807458 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os05g27170 |
| SBS | Chr5 | 21840400 | A-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os05g37340 |
| SBS | Chr6 | 13480440 | A-T | HOMO | INTRON | |
| SBS | Chr6 | 24316499 | T-C | HOMO | INTERGENIC | |
| SBS | Chr7 | 10682045 | A-T | HOMO | INTRON | |
| SBS | Chr7 | 10700544 | A-C | HOMO | INTERGENIC | |
| SBS | Chr7 | 19545702 | T-C | HOMO | UTR_3_PRIME | |
| SBS | Chr8 | 17821455 | A-C | HOMO | INTERGENIC | |
| SBS | Chr8 | 21730299 | A-C | HET | INTERGENIC | |
| SBS | Chr8 | 3411454 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 6395685 | C-T | HOMO | INTERGENIC | |
| SBS | Chr8 | 8777938 | G-A | HOMO | INTERGENIC | |
| SBS | Chr9 | 481349 | C-T | HOMO | INTRON |
Deletions: 45
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr8 | 1585050 | 1585052 | 2 | |
| Deletion | Chr8 | 2370312 | 2370314 | 2 | |
| Deletion | Chr4 | 3918642 | 3918643 | 1 | |
| Deletion | Chr10 | 4320795 | 4320849 | 54 | |
| Deletion | Chr8 | 6086096 | 6086100 | 4 | LOC_Os08g10380 |
| Deletion | Chr10 | 6874019 | 6874048 | 29 | |
| Deletion | Chr11 | 7452808 | 7452809 | 1 | |
| Deletion | Chr1 | 7900970 | 7900977 | 7 | |
| Deletion | Chr10 | 8315449 | 8315458 | 9 | LOC_Os10g16670 |
| Deletion | Chr12 | 9114774 | 9114804 | 30 | |
| Deletion | Chr3 | 9438348 | 9438349 | 1 | |
| Deletion | Chr6 | 9935092 | 9935110 | 18 | |
| Deletion | Chr12 | 11256496 | 11256499 | 3 | |
| Deletion | Chr5 | 12103903 | 12103904 | 1 | |
| Deletion | Chr1 | 12424397 | 12424405 | 8 | |
| Deletion | Chr11 | 12489094 | 12489150 | 56 | |
| Deletion | Chr8 | 12631918 | 12631947 | 29 | |
| Deletion | Chr3 | 12881950 | 12881952 | 2 | |
| Deletion | Chr10 | 13818458 | 13818459 | 1 | |
| Deletion | Chr1 | 13855483 | 13855494 | 11 | |
| Deletion | Chr7 | 14722465 | 14722477 | 12 | |
| Deletion | Chr6 | 15526689 | 15526690 | 1 | |
| Deletion | Chr9 | 16214876 | 16214878 | 2 | |
| Deletion | Chr4 | 16977819 | 16977823 | 4 | |
| Deletion | Chr11 | 17115072 | 17115073 | 1 | |
| Deletion | Chr7 | 19591289 | 19591291 | 2 | |
| Deletion | Chr2 | 20119570 | 20119577 | 7 | |
| Deletion | Chr2 | 20527910 | 20527926 | 16 | |
| Deletion | Chr11 | 21413840 | 21413841 | 1 | |
| Deletion | Chr4 | 22319899 | 22319901 | 2 | |
| Deletion | Chr2 | 23723338 | 23723352 | 14 | |
| Deletion | Chr2 | 24816985 | 24817002 | 17 | LOC_Os02g41450 |
| Deletion | Chr3 | 25350185 | 25350189 | 4 | |
| Deletion | Chr6 | 25805318 | 25805320 | 2 | |
| Deletion | Chr3 | 25953552 | 25953587 | 35 | |
| Deletion | Chr3 | 27267001 | 27306000 | 38999 | 8 |
| Deletion | Chr7 | 27861523 | 27861573 | 50 | |
| Deletion | Chr4 | 27897158 | 27897159 | 1 | |
| Deletion | Chr11 | 28977961 | 28977962 | 1 | |
| Deletion | Chr2 | 30205081 | 30205087 | 6 | |
| Deletion | Chr2 | 33162378 | 33162380 | 2 | |
| Deletion | Chr3 | 33638441 | 33638445 | 4 | |
| Deletion | Chr3 | 33746163 | 33746173 | 10 | |
| Deletion | Chr3 | 34155012 | 34155024 | 12 | LOC_Os03g60050 |
| Deletion | Chr3 | 34827426 | 34827428 | 2 |
Insertions: 11
No Inversion
No Translocation
