Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2729-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2729-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 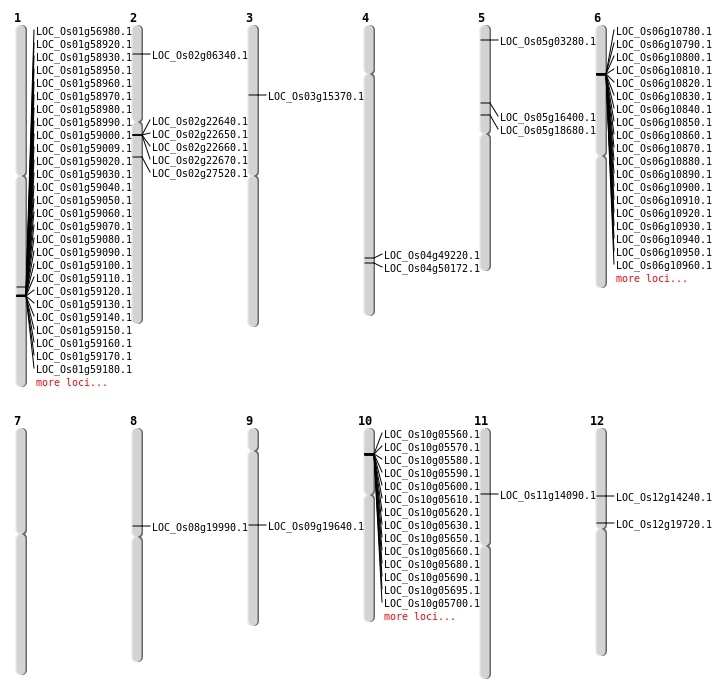
|
| Variant Information |
|---|
Single base substitutions: 115
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 21632839 | G-A | HET | ||
| SBS | Chr1 | 24636194 | T-C | Homo | ||
| SBS | Chr1 | 24636194 | T-C | HOMO | INTRON | |
| SBS | Chr1 | 28962461 | C-T | Homo | ||
| SBS | Chr10 | 11323703 | T-C | HET | ||
| SBS | Chr10 | 11323703 | T-C | HET | INTERGENIC | |
| SBS | Chr10 | 11551343 | G-A | HET | ||
| SBS | Chr10 | 11551343 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 15077092 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g28930 |
| SBS | Chr10 | 20949319 | C-G | HET | ||
| SBS | Chr10 | 4712092 | C-G | HET | ||
| SBS | Chr10 | 4712092 | C-G | HET | INTRON | |
| SBS | Chr10 | 6161604 | A-G | HET | ||
| SBS | Chr10 | 7470337 | G-A | HET | ||
| SBS | Chr10 | 7470337 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 7485821 | C-T | HET | ||
| SBS | Chr11 | 15677527 | T-A | Homo | ||
| SBS | Chr11 | 15677527 | T-A | HOMO | INTRON | |
| SBS | Chr11 | 19443309 | C-T | HET | ||
| SBS | Chr11 | 25706488 | T-A | HET | ||
| SBS | Chr11 | 25706488 | T-A | HET | INTERGENIC | |
| SBS | Chr11 | 6458726 | T-C | HET | ||
| SBS | Chr11 | 969648 | A-T | HET | ||
| SBS | Chr11 | 969648 | A-T | HET | INTERGENIC | |
| SBS | Chr11 | 9861882 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 11356787 | G-A | HET | ||
| SBS | Chr12 | 11356787 | G-A | HET | UTR_3_PRIME | |
| SBS | Chr12 | 11488441 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os12g19720 |
| SBS | Chr12 | 17178116 | G-A | HET | ||
| SBS | Chr12 | 8096864 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os12g14240 |
| SBS | Chr12 | 8324054 | G-A | HET | ||
| SBS | Chr12 | 9855012 | T-A | HET | ||
| SBS | Chr2 | 10062556 | A-T | HET | INTERGENIC | |
| SBS | Chr2 | 14012277 | G-T | HET | ||
| SBS | Chr2 | 14012277 | G-T | HET | INTRON | |
| SBS | Chr2 | 16294762 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g27520 |
| SBS | Chr2 | 3167463 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g06340 |
| SBS | Chr2 | 32058510 | T-A | HET | ||
| SBS | Chr3 | 11675304 | A-C | Homo | ||
| SBS | Chr3 | 11675304 | A-C | HOMO | INTRON | |
| SBS | Chr3 | 11675305 | C-A | Homo | ||
| SBS | Chr3 | 11675305 | C-A | HOMO | INTRON | |
| SBS | Chr3 | 19789352 | G-T | HET | ||
| SBS | Chr3 | 25761735 | C-T | Homo | ||
| SBS | Chr3 | 25761735 | C-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 34027507 | G-A | HET | ||
| SBS | Chr3 | 34027507 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 35288532 | C-A | HET | ||
| SBS | Chr4 | 14547660 | G-A | Homo | ||
| SBS | Chr4 | 15468028 | C-T | HET | ||
| SBS | Chr4 | 19406370 | C-T | HET | ||
| SBS | Chr4 | 19406370 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 22772961 | C-A | HET | ||
| SBS | Chr4 | 22772961 | C-A | HET | INTERGENIC | |
| SBS | Chr4 | 23040975 | T-G | HET | ||
| SBS | Chr4 | 23040975 | T-G | HET | INTRON | |
| SBS | Chr4 | 25729965 | A-T | HET | ||
| SBS | Chr4 | 25729965 | A-T | HET | INTRON | |
| SBS | Chr4 | 29933990 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g50172 |
| SBS | Chr4 | 29933991 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g50172 |
| SBS | Chr5 | 10823678 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g18680 |
| SBS | Chr5 | 26344327 | A-C | HET | ||
| SBS | Chr5 | 26344327 | A-C | HET | INTERGENIC | |
| SBS | Chr5 | 3579783 | C-T | Homo | ||
| SBS | Chr5 | 5317001 | A-T | HET | ||
| SBS | Chr5 | 5317001 | A-T | HET | INTERGENIC | |
| SBS | Chr5 | 6141914 | T-A | HET | ||
| SBS | Chr5 | 6141914 | T-A | HET | INTERGENIC | |
| SBS | Chr5 | 6141915 | C-A | HET | ||
| SBS | Chr5 | 6141915 | C-A | HET | INTERGENIC | |
| SBS | Chr5 | 9301646 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g16400 |
| SBS | Chr5 | 9989085 | G-T | HET | ||
| SBS | Chr6 | 15044528 | C-T | HET | ||
| SBS | Chr6 | 22386679 | T-C | HET | ||
| SBS | Chr6 | 27733178 | A-T | HET | ||
| SBS | Chr6 | 27733178 | A-T | HET | INTRON | |
| SBS | Chr6 | 7782657 | T-A | Homo | ||
| SBS | Chr6 | 7782657 | T-A | HOMO | INTRON | |
| SBS | Chr7 | 16525832 | T-C | Homo | ||
| SBS | Chr7 | 16525832 | T-C | HOMO | INTERGENIC | |
| SBS | Chr7 | 18921814 | G-T | Homo | ||
| SBS | Chr7 | 18921814 | G-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 19766393 | C-T | Homo | ||
| SBS | Chr7 | 19766393 | C-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr7 | 20321499 | G-A | HET | ||
| SBS | Chr7 | 26122265 | G-A | Homo | ||
| SBS | Chr7 | 26122265 | G-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 7937477 | T-A | HET | ||
| SBS | Chr7 | 7937477 | T-A | HET | INTRON | |
| SBS | Chr7 | 7937478 | G-A | HET | ||
| SBS | Chr7 | 7937478 | G-A | HET | INTRON | |
| SBS | Chr7 | 7937479 | G-A | HET | INTRON | |
| SBS | Chr7 | 7938125 | C-A | HET | ||
| SBS | Chr7 | 7938125 | C-A | HET | INTERGENIC | |
| SBS | Chr7 | 7938126 | T-A | HET | ||
| SBS | Chr7 | 7938126 | T-A | HET | INTERGENIC | |
| SBS | Chr8 | 11943277 | G-A | HET | ||
| SBS | Chr8 | 11972147 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os08g19990 |
| SBS | Chr8 | 17063860 | G-T | HET | ||
| SBS | Chr8 | 17063860 | G-T | HET | INTRON | |
| SBS | Chr9 | 17758474 | C-T | HET | ||
| SBS | Chr9 | 20207254 | G-A | Homo | ||
| SBS | Chr9 | 20207254 | G-A | HOMO | INTERGENIC | |
| SBS | Chr9 | 4076048 | C-T | HET | ||
| SBS | Chr9 | 4214406 | T-A | HET | ||
| SBS | Chr9 | 4214406 | T-A | HET | INTERGENIC | |
| SBS | Chr9 | 5175425 | T-C | HET | ||
| SBS | Chr9 | 5175425 | T-C | HET | INTERGENIC | |
| SBS | Chr9 | 5640990 | C-T | HET | ||
| SBS | Chr9 | 7068746 | G-T | HET | ||
| SBS | Chr9 | 8371633 | G-A | Homo | ||
| SBS | Chr9 | 8371633 | G-A | HOMO | INTERGENIC | |
| SBS | Chr9 | 8908129 | C-T | HET | ||
| SBS | Chr9 | 9711270 | T-C | Homo | ||
| SBS | Chr9 | 9711270 | T-C | HOMO | INTRON |
Deletions: 43
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr10 | 2760001 | 2947000 | 186999 | 50 |
| Deletion | Chr10 | 2960001 | 2978000 | 17999 | 4 |
| Deletion | Chr10 | 2961001 | 2978000 | 16999 | 3 |
| Deletion | Chr10 | 2981001 | 3062000 | 80999 | 12 |
| Deletion | Chr10 | 2982001 | 3059000 | 76999 | 11 |
| Deletion | Chr6 | 4329483 | 4329581 | 98 | |
| Deletion | Chr6 | 5627001 | 5893000 | 265999 | 45 |
| Deletion | Chr6 | 5631001 | 5882000 | 250999 | 43 |
| Deletion | Chr1 | 5726034 | 5726036 | 2 | |
| Deletion | Chr9 | 5770956 | 5770966 | 10 | |
| Deletion | Chr6 | 5895001 | 5965000 | 69999 | 41 |
| Deletion | Chr6 | 5970001 | 6109000 | 138999 | 17 |
| Deletion | Chr7 | 6021662 | 6021673 | 11 | |
| Deletion | Chr10 | 6235035 | 6235047 | 12 | |
| Deletion | Chr8 | 7297314 | 7297315 | 1 | |
| Deletion | Chr8 | 7901863 | 7901867 | 4 | |
| Deletion | Chr9 | 8170450 | 8170455 | 5 | 2 |
| Deletion | Chr11 | 9517333 | 9517339 | 6 | 2 |
| Deletion | Chr1 | 10347968 | 10347970 | 2 | |
| Deletion | Chr9 | 11739692 | 11739696 | 4 | 2 |
| Deletion | Chr6 | 12545870 | 12545871 | 1 | LOC_Os06g21700 |
| Deletion | Chr10 | 12635969 | 12635978 | 9 | LOC_Os10g24620 |
| Deletion | Chr3 | 12858226 | 12858231 | 5 | 2 |
| Deletion | Chr2 | 13491001 | 13504000 | 12999 | 4 |
| Deletion | Chr8 | 14189620 | 14189624 | 4 | 2 |
| Deletion | Chr1 | 16975172 | 16975173 | 1 | |
| Deletion | Chr4 | 18468602 | 18468603 | 1 | |
| Deletion | Chr1 | 19679082 | 19679088 | 6 | |
| Deletion | Chr12 | 23417167 | 23417171 | 4 | 2 |
| Deletion | Chr6 | 24395001 | 24395009 | 8 | |
| Deletion | Chr12 | 24816125 | 24816133 | 8 | 2 |
| Deletion | Chr7 | 26123544 | 26123612 | 68 | |
| Deletion | Chr11 | 26982412 | 26982413 | 1 | |
| Deletion | Chr8 | 27625588 | 27625590 | 2 | |
| Deletion | Chr1 | 28209328 | 28209330 | 2 | |
| Deletion | Chr4 | 29349180 | 29349182 | 2 | LOC_Os04g49220 |
| Deletion | Chr1 | 34045001 | 34369000 | 323999 | 48 |
| Deletion | Chr1 | 34053001 | 34338000 | 284999 | 44 |
| Deletion | Chr1 | 34270916 | 34270917 | 1 | |
| Deletion | Chr1 | 34347001 | 34368000 | 20999 | 3 |
| Deletion | Chr1 | 35296180 | 35296184 | 4 | |
| Deletion | Chr1 | 36765001 | 36780000 | 14999 | 4 |
| Deletion | Chr1 | 37879178 | 37879179 | 1 |
Insertions: 19
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 10779952 | 10779961 | 10 | |
| Insertion | Chr1 | 14045450 | 14045457 | 8 | |
| Insertion | Chr1 | 36793288 | 36793297 | 10 | |
| Insertion | Chr12 | 20273415 | 20273416 | 2 | |
| Insertion | Chr3 | 21326837 | 21326837 | 1 | |
| Insertion | Chr3 | 28982499 | 28982500 | 2 | 2 |
| Insertion | Chr3 | 28982499 | 28982500 | 2 | 2 |
| Insertion | Chr4 | 11052243 | 11052244 | 2 | |
| Insertion | Chr4 | 16977819 | 16977822 | 4 | |
| Insertion | Chr4 | 22771401 | 22771412 | 12 | |
| Insertion | Chr5 | 1329961 | 1329961 | 1 | LOC_Os05g03280 |
| Insertion | Chr5 | 8256416 | 8256416 | 1 | |
| Insertion | Chr6 | 11000810 | 11000810 | 1 | |
| Insertion | Chr6 | 22472065 | 22472065 | 1 | |
| Insertion | Chr7 | 2572048 | 2572048 | 1 | |
| Insertion | Chr7 | 4427071 | 4427072 | 2 | |
| Insertion | Chr7 | 5731836 | 5731836 | 1 | |
| Insertion | Chr8 | 22509329 | 22509329 | 1 | |
| Insertion | Chr9 | 987222 | 987231 | 10 |
Inversions: 6
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr10 | 2761404 | 2762165 | LOC_Os10g05560 |
| Inversion | Chr10 | 2761648 | 2762158 | LOC_Os10g05560 |
| Inversion | Chr10 | 3058948 | 4031510 | |
| Inversion | Chr10 | 5522056 | 5522323 | LOC_Os10g10110 |
| Inversion | Chr6 | 6110601 | 6713679 | |
| Inversion | Chr1 | 32924190 | 32924346 | LOC_Os01g56980 |
Translocations: 12
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 604732 | Chr4 | 3063060 | |
| Translocation | Chr11 | 6135697 | Chr1 | 35969205 | |
| Translocation | Chr11 | 7846350 | Chr6 | 16078486 | 2 |
| Translocation | Chr5 | 7917874 | Chr3 | 8412207 | 2 |
| Translocation | Chr9 | 8226708 | Chr2 | 11631204 | |
| Translocation | Chr12 | 19144011 | Chr5 | 18325140 | |
| Translocation | Chr11 | 19748567 | Chr6 | 5893166 | |
| Translocation | Chr6 | 22189392 | Chr4 | 24563559 | |
| Translocation | Chr11 | 23368404 | Chr5 | 13251372 | |
| Translocation | Chr11 | 25694182 | Chr10 | 9261636 | |
| Translocation | Chr11 | 25694526 | Chr10 | 4031488 | |
| Translocation | Chr3 | 31068314 | Chr1 | 16252169 |
