Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2735-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2735-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 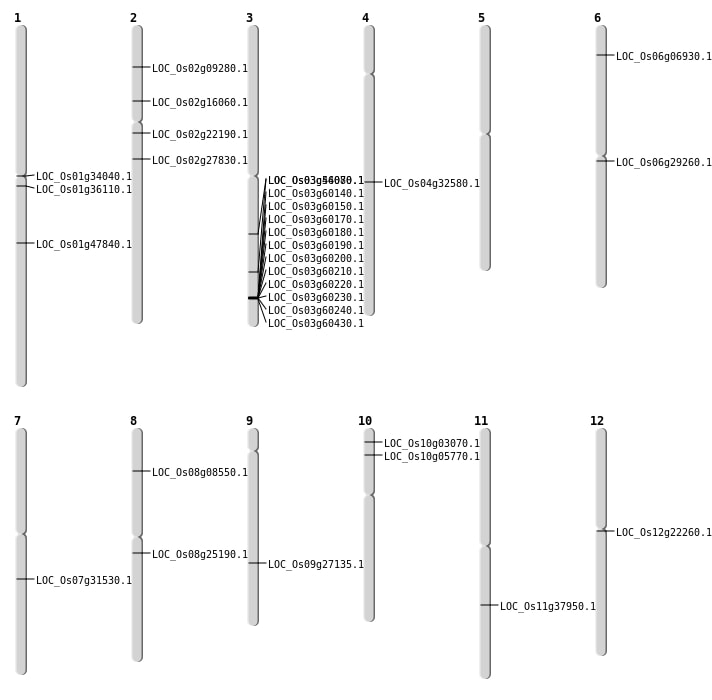
|
| Variant Information |
|---|
Single base substitutions: 60
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 27387925 | C-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os01g47840 |
| SBS | Chr1 | 27387925 | C-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os01g47840 |
| SBS | Chr1 | 616894 | C-T | HET | ||
| SBS | Chr1 | 8272420 | T-G | HET | ||
| SBS | Chr1 | 8272420 | T-G | HET | UTR_3_PRIME | |
| SBS | Chr1 | 9168536 | G-A | Homo | ||
| SBS | Chr1 | 9168536 | G-A | HOMO | INTRON | |
| SBS | Chr10 | 1280514 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os10g03070 |
| SBS | Chr10 | 4415597 | C-T | HET | ||
| SBS | Chr10 | 6931317 | A-T | HET | ||
| SBS | Chr10 | 6931317 | A-T | HET | INTERGENIC | |
| SBS | Chr11 | 5592997 | C-T | Homo | ||
| SBS | Chr11 | 5592997 | C-T | HOMO | UTR_3_PRIME | |
| SBS | Chr12 | 17047000 | C-G | HET | ||
| SBS | Chr12 | 17047000 | C-G | HET | INTRON | |
| SBS | Chr12 | 9201439 | G-T | HET | ||
| SBS | Chr2 | 32872840 | G-A | HET | ||
| SBS | Chr2 | 32872840 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 35082389 | C-T | HET | ||
| SBS | Chr2 | 35082389 | C-T | HET | INTRON | |
| SBS | Chr2 | 4780196 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g09280 |
| SBS | Chr3 | 11934805 | G-C | Homo | ||
| SBS | Chr3 | 11934805 | G-C | HOMO | INTERGENIC | |
| SBS | Chr3 | 21049440 | G-A | HET | ||
| SBS | Chr3 | 21049440 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 21049441 | C-A | HET | ||
| SBS | Chr3 | 21049441 | C-A | HET | INTERGENIC | |
| SBS | Chr3 | 22719043 | C-A | HET | ||
| SBS | Chr3 | 22719043 | C-A | HET | INTERGENIC | |
| SBS | Chr3 | 22719044 | T-A | HET | ||
| SBS | Chr3 | 22719044 | T-A | HET | INTERGENIC | |
| SBS | Chr3 | 28326449 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 5625085 | T-C | Homo | ||
| SBS | Chr3 | 5625085 | T-C | HOMO | INTERGENIC | |
| SBS | Chr3 | 8739209 | C-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr4 | 11967 | C-T | HET | ||
| SBS | Chr4 | 19611566 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g32580 |
| SBS | Chr4 | 28193306 | C-A | Homo | ||
| SBS | Chr4 | 28193306 | C-A | HOMO | INTRON | |
| SBS | Chr5 | 19407161 | G-T | HET | ||
| SBS | Chr5 | 19407161 | G-T | HET | INTERGENIC | |
| SBS | Chr6 | 11976900 | T-C | HET | ||
| SBS | Chr6 | 11976900 | T-C | HET | INTERGENIC | |
| SBS | Chr6 | 11976901 | A-T | HET | ||
| SBS | Chr6 | 11976901 | A-T | HET | INTERGENIC | |
| SBS | Chr6 | 11976902 | C-T | HET | ||
| SBS | Chr6 | 11976902 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 12769637 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 16715294 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os06g29260 |
| SBS | Chr6 | 31190382 | T-G | Homo | ||
| SBS | Chr6 | 31190382 | T-G | HOMO | INTERGENIC | |
| SBS | Chr6 | 8735655 | G-T | Homo | ||
| SBS | Chr6 | 8735655 | G-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 9699199 | C-A | HET | ||
| SBS | Chr8 | 12265008 | G-C | HET | ||
| SBS | Chr8 | 12265008 | G-C | HET | INTERGENIC | |
| SBS | Chr8 | 26986863 | A-C | HET | ||
| SBS | Chr8 | 26986863 | A-C | HET | INTERGENIC | |
| SBS | Chr8 | 7779656 | C-T | HET | ||
| SBS | Chr9 | 16501564 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os09g27135 |
Deletions: 51
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr3 | 133457 | 133458 | 1 | |
| Deletion | Chr10 | 518604 | 518605 | 1 | |
| Deletion | Chr1 | 992250 | 992295 | 45 | |
| Deletion | Chr8 | 1911419 | 1911421 | 2 | |
| Deletion | Chr4 | 1937797 | 1937798 | 1 | |
| Deletion | Chr5 | 2405200 | 2405201 | 1 | |
| Deletion | Chr1 | 2906206 | 2906207 | 1 | |
| Deletion | Chr6 | 3287783 | 3287789 | 6 | 2 |
| Deletion | Chr2 | 3819903 | 3819904 | 1 | |
| Deletion | Chr8 | 4252392 | 4252457 | 65 | |
| Deletion | Chr9 | 4759101 | 4759106 | 5 | |
| Deletion | Chr4 | 5306107 | 5306116 | 9 | |
| Deletion | Chr10 | 5857326 | 5857327 | 1 | |
| Deletion | Chr8 | 7283058 | 7283059 | 1 | |
| Deletion | Chr10 | 8728114 | 8728115 | 1 | |
| Deletion | Chr12 | 8895880 | 8895882 | 2 | |
| Deletion | Chr5 | 9565373 | 9565374 | 1 | |
| Deletion | Chr5 | 9908248 | 9908249 | 1 | |
| Deletion | Chr8 | 10456913 | 10456941 | 28 | |
| Deletion | Chr1 | 10884004 | 10884006 | 2 | |
| Deletion | Chr9 | 11015974 | 11015980 | 6 | 2 |
| Deletion | Chr7 | 11447635 | 11447636 | 1 | |
| Deletion | Chr12 | 12035144 | 12035309 | 165 | |
| Deletion | Chr7 | 12166680 | 12166681 | 1 | |
| Deletion | Chr7 | 12169778 | 12169779 | 1 | |
| Deletion | Chr3 | 12206509 | 12206510 | 1 | |
| Deletion | Chr3 | 12206713 | 12206714 | 1 | |
| Deletion | Chr8 | 12584371 | 12584372 | 1 | |
| Deletion | Chr4 | 12686839 | 12686840 | 1 | |
| Deletion | Chr2 | 13051627 | 13051629 | 2 | |
| Deletion | Chr6 | 14040088 | 14040089 | 1 | |
| Deletion | Chr4 | 14619868 | 14619885 | 17 | |
| Deletion | Chr8 | 15304149 | 15304150 | 1 | LOC_Os08g25190 |
| Deletion | Chr10 | 15534467 | 15534489 | 22 | |
| Deletion | Chr7 | 17305022 | 17305023 | 1 | |
| Deletion | Chr11 | 17738876 | 17738877 | 1 | |
| Deletion | Chr6 | 19641970 | 19641971 | 1 | |
| Deletion | Chr2 | 20070677 | 20070683 | 6 | |
| Deletion | Chr5 | 22014692 | 22014693 | 1 | |
| Deletion | Chr10 | 22621280 | 22621282 | 2 | |
| Deletion | Chr1 | 23494088 | 23494093 | 5 | |
| Deletion | Chr3 | 26061955 | 26061956 | 1 | LOC_Os03g46080 |
| Deletion | Chr8 | 26672356 | 26672357 | 1 | |
| Deletion | Chr8 | 26736967 | 26736973 | 6 | |
| Deletion | Chr11 | 26895145 | 26895146 | 1 | |
| Deletion | Chr2 | 28384425 | 28384446 | 21 | |
| Deletion | Chr4 | 29551232 | 29551348 | 116 | |
| Deletion | Chr3 | 34199001 | 34262000 | 62999 | 10 |
| Deletion | Chr3 | 34201001 | 34261000 | 59999 | 10 |
| Deletion | Chr1 | 36921033 | 36921034 | 1 | |
| Deletion | Chr1 | 37206411 | 37206429 | 18 |
Insertions: 29
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 11281091 | 11281091 | 1 | |
| Insertion | Chr1 | 16220855 | 16220855 | 1 | |
| Insertion | Chr1 | 19621410 | 19621410 | 1 | |
| Insertion | Chr1 | 23401001 | 23401006 | 6 | |
| Insertion | Chr1 | 26678442 | 26678442 | 1 | |
| Insertion | Chr1 | 33860431 | 33860431 | 1 | |
| Insertion | Chr1 | 5763183 | 5763188 | 6 | |
| Insertion | Chr10 | 18877063 | 18877074 | 12 | |
| Insertion | Chr10 | 20977113 | 20977113 | 1 | |
| Insertion | Chr12 | 18817390 | 18817390 | 1 | |
| Insertion | Chr2 | 21223671 | 21223671 | 1 | |
| Insertion | Chr2 | 23353062 | 23353062 | 1 | |
| Insertion | Chr2 | 5699213 | 5699218 | 6 | |
| Insertion | Chr2 | 9139264 | 9139264 | 1 | LOC_Os02g16060 |
| Insertion | Chr4 | 26597673 | 26597673 | 1 | |
| Insertion | Chr4 | 28033918 | 28033918 | 1 | |
| Insertion | Chr5 | 13212615 | 13212615 | 1 | |
| Insertion | Chr5 | 17503644 | 17503649 | 6 | |
| Insertion | Chr5 | 29750609 | 29750616 | 8 | |
| Insertion | Chr6 | 11110065 | 11110065 | 1 | |
| Insertion | Chr6 | 19967524 | 19967526 | 3 | |
| Insertion | Chr7 | 26050686 | 26050686 | 1 | |
| Insertion | Chr8 | 10714504 | 10714505 | 2 | |
| Insertion | Chr8 | 17547260 | 17547260 | 1 | |
| Insertion | Chr8 | 24481451 | 24481453 | 3 | |
| Insertion | Chr8 | 4932614 | 4932614 | 1 | LOC_Os08g08550 |
| Insertion | Chr8 | 5192175 | 5192177 | 3 | 2 |
| Insertion | Chr8 | 5192175 | 5192181 | 7 | 2 |
| Insertion | Chr9 | 10011417 | 10011417 | 1 |
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr11 | 959677 | 963747 |
Translocations: 35
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 728649 | Chr2 | 13225552 | 2 |
| Translocation | Chr7 | 1503924 | Chr4 | 3249683 | |
| Translocation | Chr8 | 2694146 | Chr1 | 16146323 | |
| Translocation | Chr7 | 3694378 | Chr4 | 3213554 | |
| Translocation | Chr7 | 5440325 | Chr6 | 7372684 | |
| Translocation | Chr10 | 6538384 | Chr4 | 9746190 | |
| Translocation | Chr9 | 7218440 | Chr2 | 4247059 | |
| Translocation | Chr9 | 7228101 | Chr6 | 7358335 | |
| Translocation | Chr8 | 7820676 | Chr1 | 8343013 | |
| Translocation | Chr12 | 11058664 | Chr8 | 15320629 | |
| Translocation | Chr12 | 11861800 | Chr11 | 1474966 | |
| Translocation | Chr12 | 12547956 | Chr10 | 18187587 | LOC_Os12g22260 |
| Translocation | Chr6 | 14459837 | Chr4 | 8661879 | |
| Translocation | Chr11 | 16827494 | Chr10 | 2906715 | LOC_Os10g05770 |
| Translocation | Chr8 | 18074316 | Chr1 | 18737722 | LOC_Os01g34040 |
| Translocation | Chr11 | 18177016 | Chr8 | 17758093 | |
| Translocation | Chr2 | 18249663 | Chr1 | 15324159 | |
| Translocation | Chr5 | 18271745 | Chr4 | 28907054 | |
| Translocation | Chr7 | 18715623 | Chr1 | 2892092 | LOC_Os07g31530 |
| Translocation | Chr4 | 19146715 | Chr3 | 17621955 | |
| Translocation | Chr6 | 19268947 | Chr1 | 15459835 | |
| Translocation | Chr8 | 19690453 | Chr6 | 13904217 | |
| Translocation | Chr12 | 20096544 | Chr6 | 23395012 | |
| Translocation | Chr8 | 20135621 | Chr1 | 2999829 | |
| Translocation | Chr10 | 20165813 | Chr4 | 31852132 | |
| Translocation | Chr6 | 20520242 | Chr1 | 878755 | |
| Translocation | Chr12 | 21494618 | Chr11 | 17511549 | |
| Translocation | Chr12 | 22003598 | Chr2 | 33566140 | |
| Translocation | Chr9 | 22299529 | Chr2 | 21647280 | |
| Translocation | Chr11 | 22502094 | Chr10 | 3012238 | LOC_Os11g37950 |
| Translocation | Chr6 | 23802805 | Chr3 | 4986014 | |
| Translocation | Chr12 | 26916650 | Chr2 | 17337443 | |
| Translocation | Chr12 | 27085284 | Chr11 | 18154422 | |
| Translocation | Chr3 | 30999477 | Chr1 | 19980757 | 2 |
| Translocation | Chr3 | 35300731 | Chr2 | 16481527 | 2 |
