Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2749-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2749-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 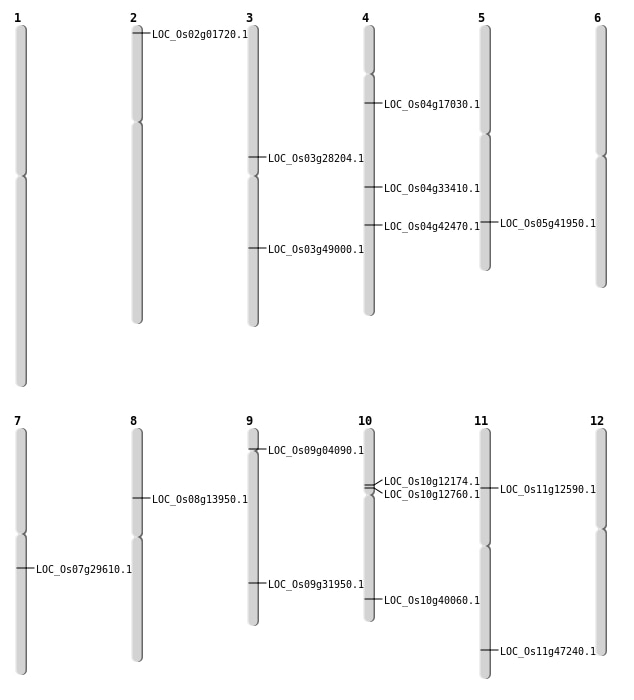
|
| Variant Information |
|---|
Single base substitutions: 55
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 24292437 | C-T | Homo | ||
| SBS | Chr1 | 37539717 | G-A | HET | ||
| SBS | Chr10 | 12417776 | C-T | HET | ||
| SBS | Chr10 | 21456170 | T-G | HET | NON_SYNONYMOUS_CODING | LOC_Os10g40060 |
| SBS | Chr10 | 22830732 | T-C | HET | ||
| SBS | Chr10 | 6727770 | G-A | HET | ||
| SBS | Chr10 | 6790383 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os10g12174 |
| SBS | Chr10 | 7125298 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os10g12760 |
| SBS | Chr10 | 9694365 | G-A | HET | ||
| SBS | Chr11 | 1522543 | C-T | HET | ||
| SBS | Chr11 | 19953774 | C-A | HET | ||
| SBS | Chr11 | 20783228 | T-A | Homo | ||
| SBS | Chr11 | 24298717 | T-A | HET | ||
| SBS | Chr11 | 4427695 | T-G | HET | ||
| SBS | Chr11 | 9382366 | C-A | HET | ||
| SBS | Chr12 | 13307186 | T-C | HET | ||
| SBS | Chr12 | 19959223 | C-G | HET | ||
| SBS | Chr12 | 21898 | C-T | HET | ||
| SBS | Chr12 | 4848630 | G-A | HET | ||
| SBS | Chr2 | 1334260 | G-A | HET | ||
| SBS | Chr2 | 1849852 | A-T | Homo | ||
| SBS | Chr2 | 23796244 | C-T | Homo | ||
| SBS | Chr2 | 27517627 | G-C | HET | ||
| SBS | Chr2 | 5990261 | C-A | HET | ||
| SBS | Chr3 | 16236892 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os03g28204 |
| SBS | Chr3 | 31164605 | G-A | HET | ||
| SBS | Chr3 | 32943286 | G-T | HET | ||
| SBS | Chr3 | 4743070 | C-A | Homo | ||
| SBS | Chr4 | 13888214 | G-A | HET | ||
| SBS | Chr4 | 15431933 | C-A | HET | ||
| SBS | Chr4 | 18283833 | A-C | Homo | ||
| SBS | Chr4 | 27441010 | G-T | HET | ||
| SBS | Chr4 | 28222363 | A-G | HET | ||
| SBS | Chr4 | 34520291 | A-T | HET | ||
| SBS | Chr4 | 4104665 | C-T | Homo | ||
| SBS | Chr4 | 6738485 | G-T | HET | ||
| SBS | Chr4 | 9438294 | G-T | HET | ||
| SBS | Chr5 | 14444677 | A-T | Homo | ||
| SBS | Chr5 | 15984961 | G-T | Homo | ||
| SBS | Chr5 | 18122488 | T-C | HET | ||
| SBS | Chr5 | 20860796 | C-T | HET | ||
| SBS | Chr5 | 8555260 | G-A | HET | ||
| SBS | Chr6 | 17253975 | C-G | HET | ||
| SBS | Chr6 | 17819640 | C-A | HET | ||
| SBS | Chr7 | 9465809 | A-G | HET | ||
| SBS | Chr7 | 9465810 | G-A | HET | ||
| SBS | Chr8 | 12128455 | A-C | HET | ||
| SBS | Chr8 | 12128456 | G-T | HET | ||
| SBS | Chr8 | 19846753 | T-C | Homo | ||
| SBS | Chr8 | 8359394 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g13950 |
| SBS | Chr8 | 8772992 | G-A | HET | ||
| SBS | Chr9 | 13551491 | C-A | HET | ||
| SBS | Chr9 | 19051274 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os09g31950 |
| SBS | Chr9 | 19350610 | A-G | HET | ||
| SBS | Chr9 | 2119509 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os09g04090 |
Deletions: 7
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr1 | 8852985 | 8852986 | 1 | |
| Deletion | Chr11 | 16553159 | 16553160 | 1 | |
| Deletion | Chr4 | 20234533 | 20234535 | 2 | LOC_Os04g33410 |
| Deletion | Chr2 | 21085982 | 21085994 | 12 | |
| Deletion | Chr3 | 25659549 | 25659553 | 4 | |
| Deletion | Chr3 | 27595606 | 27595610 | 4 | |
| Deletion | Chr3 | 27909633 | 27909654 | 21 | LOC_Os03g49000 |
Insertions: 10
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 2919716 | 2919716 | 1 | |
| Insertion | Chr1 | 29577487 | 29577488 | 2 | |
| Insertion | Chr1 | 8612246 | 8612247 | 2 | |
| Insertion | Chr4 | 4268572 | 4268572 | 1 | |
| Insertion | Chr4 | 9327943 | 9327943 | 1 | LOC_Os04g17030 |
| Insertion | Chr6 | 17092357 | 17092357 | 1 | |
| Insertion | Chr7 | 1416569 | 1416570 | 2 | |
| Insertion | Chr8 | 17910649 | 17910666 | 18 | |
| Insertion | Chr8 | 23317908 | 23317908 | 1 | |
| Insertion | Chr9 | 8299252 | 8299252 | 1 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr7 | 7026346 | 7150445 | |
| Inversion | Chr7 | 7026349 | 7150456 |
Translocations: 6
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 7073666 | Chr7 | 17406846 | 2 |
| Translocation | Chr10 | 15776444 | Chr2 | 397595 | 2 |
| Translocation | Chr3 | 16448154 | Chr1 | 25435244 | |
| Translocation | Chr5 | 24570204 | Chr4 | 25131128 | 2 |
| Translocation | Chr4 | 25131566 | Chr3 | 23200686 | LOC_Os04g42470 |
| Translocation | Chr11 | 28407988 | Chr2 | 13043311 | LOC_Os11g47240 |
