Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2752-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2752-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 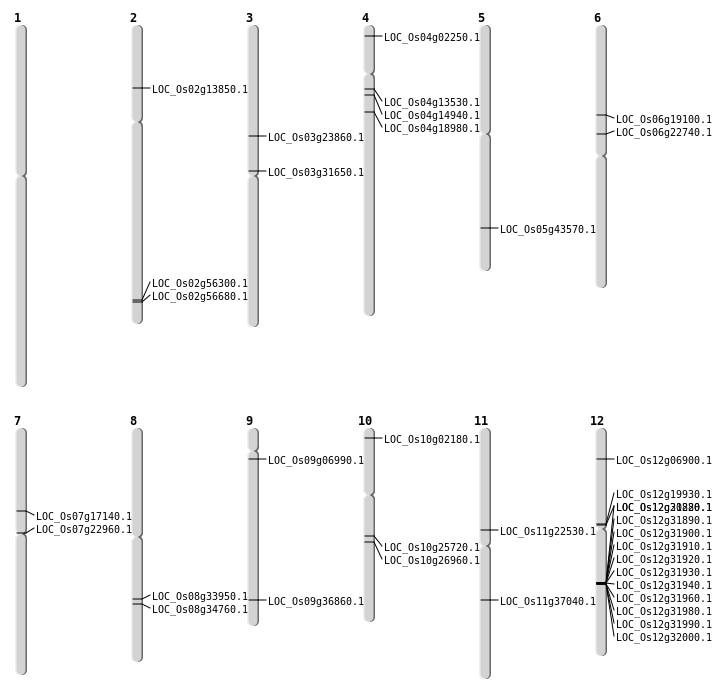
|
| Variant Information |
|---|
Single base substitutions: 86
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 11085472 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 38329087 | T-A | HET | INTERGENIC | |
| SBS | Chr1 | 5734781 | T-C | HET | ||
| SBS | Chr10 | 13322043 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g25720 |
| SBS | Chr10 | 14198418 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g26960 |
| SBS | Chr10 | 16923342 | G-A | Homo | ||
| SBS | Chr10 | 16936883 | C-A | HET | ||
| SBS | Chr10 | 19310054 | C-T | Homo | ||
| SBS | Chr10 | 268031 | T-A | HET | INTERGENIC | |
| SBS | Chr10 | 8924128 | T-C | HET | ||
| SBS | Chr11 | 13595362 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 13595363 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 14463859 | T-C | HET | INTERGENIC | |
| SBS | Chr11 | 16839493 | G-A | Homo | ||
| SBS | Chr11 | 19312246 | C-G | HET | ||
| SBS | Chr11 | 20621523 | A-G | HET | INTERGENIC | |
| SBS | Chr11 | 21862430 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g37040 |
| SBS | Chr11 | 4450055 | G-A | HET | INTRON | |
| SBS | Chr11 | 8964784 | C-A | HET | INTERGENIC | |
| SBS | Chr12 | 11613192 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g19930 |
| SBS | Chr12 | 12126480 | T-A | HET | ||
| SBS | Chr12 | 20325124 | A-T | HET | ||
| SBS | Chr12 | 2507640 | A-G | HET | ||
| SBS | Chr12 | 25807949 | G-C | HET | UTR_3_PRIME | |
| SBS | Chr12 | 2684496 | C-T | HET | ||
| SBS | Chr12 | 3358586 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g06900 |
| SBS | Chr12 | 9791407 | G-A | HET | ||
| SBS | Chr2 | 14556979 | G-C | HET | ||
| SBS | Chr2 | 20017290 | G-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 22254500 | C-G | HET | ||
| SBS | Chr2 | 35011523 | A-G | HOMO | UTR_3_PRIME | |
| SBS | Chr3 | 15157907 | G-C | Homo | ||
| SBS | Chr3 | 21198057 | T-A | HET | ||
| SBS | Chr3 | 22836943 | G-A | HET | ||
| SBS | Chr3 | 23123639 | G-A | Homo | ||
| SBS | Chr3 | 24991745 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 26074698 | A-C | HOMO | INTRON | |
| SBS | Chr3 | 2779069 | A-C | Homo | ||
| SBS | Chr3 | 35816617 | A-C | HET | ||
| SBS | Chr3 | 4658856 | A-C | Homo | ||
| SBS | Chr3 | 7067518 | T-C | HET | ||
| SBS | Chr3 | 7533718 | C-A | HET | ||
| SBS | Chr3 | 758046 | T-A | Homo | ||
| SBS | Chr3 | 9693514 | C-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 10549755 | C-T | Homo | ||
| SBS | Chr4 | 10571919 | G-A | HET | ||
| SBS | Chr4 | 11850838 | C-T | HET | ||
| SBS | Chr4 | 13282736 | A-G | HET | INTERGENIC | |
| SBS | Chr4 | 14328301 | A-G | HET | ||
| SBS | Chr4 | 14555339 | C-T | Homo | ||
| SBS | Chr4 | 34110384 | G-T | Homo | ||
| SBS | Chr4 | 3439632 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 4394064 | A-G | Homo | ||
| SBS | Chr4 | 8402055 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g14940 |
| SBS | Chr4 | 9411765 | G-A | Homo | ||
| SBS | Chr5 | 19981449 | A-G | HET | ||
| SBS | Chr5 | 2183428 | C-T | HET | ||
| SBS | Chr5 | 24686172 | T-A | HET | ||
| SBS | Chr5 | 24878722 | C-A | HET | ||
| SBS | Chr5 | 25335409 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os05g43570 |
| SBS | Chr5 | 524062 | G-C | HET | INTERGENIC | |
| SBS | Chr6 | 16543925 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 16586458 | G-A | HET | ||
| SBS | Chr6 | 1854729 | T-C | HET | INTERGENIC | |
| SBS | Chr6 | 20182219 | C-A | HET | INTERGENIC | |
| SBS | Chr6 | 22165436 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr6 | 22281600 | G-A | HET | ||
| SBS | Chr6 | 31238421 | C-T | Homo | ||
| SBS | Chr7 | 11741318 | C-T | HET | ||
| SBS | Chr7 | 11741319 | C-T | HET | ||
| SBS | Chr7 | 17845212 | T-C | HET | ||
| SBS | Chr7 | 21742057 | G-A | HET | INTRON | |
| SBS | Chr7 | 23972237 | T-A | Homo | ||
| SBS | Chr7 | 485240 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 12324413 | C-T | Homo | ||
| SBS | Chr8 | 14123077 | A-C | HET | ||
| SBS | Chr8 | 21195051 | G-A | HOMO | INTRON | |
| SBS | Chr8 | 21195053 | G-A | HOMO | INTRON | |
| SBS | Chr8 | 21257219 | T-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os08g33950 |
| SBS | Chr8 | 25452213 | C-T | HET | ||
| SBS | Chr8 | 4288197 | G-A | Homo | ||
| SBS | Chr8 | 4419035 | T-C | HET | INTERGENIC | |
| SBS | Chr8 | 4419036 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 13755872 | T-G | HET | ||
| SBS | Chr9 | 21254558 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os09g36860 |
| SBS | Chr9 | 21254559 | G-A | HET | SYNONYMOUS_CODING |
Deletions: 44
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr7 | 229034 | 229038 | 4 | |
| Deletion | Chr2 | 813864 | 813865 | 1 | |
| Deletion | Chr6 | 1732923 | 1732924 | 1 | |
| Deletion | Chr11 | 2416414 | 2416419 | 5 | |
| Deletion | Chr6 | 3089458 | 3089472 | 14 | |
| Deletion | Chr7 | 3713278 | 3713279 | 1 | |
| Deletion | Chr12 | 4055986 | 4055992 | 6 | |
| Deletion | Chr4 | 4616765 | 4616766 | 1 | |
| Deletion | Chr11 | 5596160 | 5596161 | 1 | |
| Deletion | Chr7 | 6082689 | 6082781 | 92 | |
| Deletion | Chr8 | 6760515 | 6760516 | 1 | |
| Deletion | Chr4 | 7116361 | 7116363 | 2 | |
| Deletion | Chr2 | 7509736 | 7509761 | 25 | LOC_Os02g13850 |
| Deletion | Chr7 | 9612033 | 9612049 | 16 | |
| Deletion | Chr7 | 10071021 | 10071030 | 9 | LOC_Os07g17140 |
| Deletion | Chr4 | 10543478 | 10543479 | 1 | LOC_Os04g18980 |
| Deletion | Chr6 | 10869131 | 10869139 | 8 | LOC_Os06g19100 |
| Deletion | Chr12 | 11557621 | 11557638 | 17 | |
| Deletion | Chr4 | 11939996 | 11939998 | 2 | |
| Deletion | Chr4 | 12169992 | 12170048 | 56 | |
| Deletion | Chr9 | 12626465 | 12626467 | 2 | |
| Deletion | Chr7 | 12938685 | 12938687 | 2 | LOC_Os07g22960 |
| Deletion | Chr6 | 13228457 | 13228483 | 26 | LOC_Os06g22740 |
| Deletion | Chr2 | 14327914 | 14327915 | 1 | |
| Deletion | Chr9 | 14670798 | 14670799 | 1 | |
| Deletion | Chr4 | 14967133 | 14967137 | 4 | |
| Deletion | Chr12 | 15540968 | 15540969 | 1 | |
| Deletion | Chr4 | 16073910 | 16073956 | 46 | |
| Deletion | Chr1 | 16534207 | 16534208 | 1 | |
| Deletion | Chr1 | 17941212 | 17941222 | 10 | |
| Deletion | Chr1 | 18915877 | 18915879 | 2 | |
| Deletion | Chr12 | 19188001 | 19234000 | 45999 | 6 |
| Deletion | Chr12 | 19238001 | 19287000 | 48999 | 5 |
| Deletion | Chr2 | 19685614 | 19685617 | 3 | |
| Deletion | Chr6 | 22178018 | 22178020 | 2 | |
| Deletion | Chr7 | 22686675 | 22686743 | 68 | |
| Deletion | Chr4 | 22740769 | 22740770 | 1 | |
| Deletion | Chr2 | 22902795 | 22902796 | 1 | |
| Deletion | Chr1 | 26192332 | 26192333 | 1 | |
| Deletion | Chr4 | 30479024 | 30479027 | 3 | |
| Deletion | Chr1 | 31498875 | 31498878 | 3 | |
| Deletion | Chr1 | 33853177 | 33853178 | 1 | |
| Deletion | Chr2 | 34463476 | 34463483 | 7 | LOC_Os02g56300 |
| Deletion | Chr3 | 35708142 | 35708146 | 4 |
Insertions: 14
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 21772474 | 21772476 | 3 | |
| Insertion | Chr10 | 749754 | 749754 | 1 | LOC_Os10g02180 |
| Insertion | Chr11 | 26072450 | 26072459 | 10 | 2 |
| Insertion | Chr11 | 26072450 | 26072451 | 2 | 2 |
| Insertion | Chr12 | 21997879 | 21997880 | 2 | |
| Insertion | Chr2 | 22009569 | 22009584 | 16 | |
| Insertion | Chr3 | 13380250 | 13380250 | 1 | |
| Insertion | Chr3 | 14831618 | 14831623 | 6 | |
| Insertion | Chr3 | 14987577 | 14987577 | 1 | |
| Insertion | Chr4 | 7546470 | 7546470 | 1 | LOC_Os04g13530 |
| Insertion | Chr5 | 29103569 | 29103570 | 2 | |
| Insertion | Chr8 | 8918680 | 8918685 | 6 | |
| Insertion | Chr9 | 13663140 | 13663159 | 20 | |
| Insertion | Chr9 | 18970946 | 18970961 | 16 |
Inversions: 9
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr11 | 2223307 | 2223408 | |
| Inversion | Chr5 | 3190947 | 3969039 | |
| Inversion | Chr5 | 3190954 | 3969053 | |
| Inversion | Chr3 | 18032367 | 18073228 | LOC_Os03g31650 |
| Inversion | Chr8 | 21855186 | 22366710 | LOC_Os08g34760 |
| Inversion | Chr8 | 21855200 | 22366711 | LOC_Os08g34760 |
| Inversion | Chr8 | 24138434 | 24138581 | |
| Inversion | Chr1 | 25962861 | 26492157 | |
| Inversion | Chr1 | 25962864 | 26492159 |
Translocations: 13
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 2785627 | Chr3 | 16685673 | |
| Translocation | Chr9 | 4040357 | Chr6 | 8105987 | |
| Translocation | Chr11 | 4272848 | Chr9 | 3387619 | LOC_Os09g06990 |
| Translocation | Chr3 | 4835539 | Chr2 | 31043205 | |
| Translocation | Chr4 | 5588399 | Chr1 | 25576624 | |
| Translocation | Chr3 | 11066394 | Chr2 | 29255718 | |
| Translocation | Chr12 | 11794917 | Chr1 | 14847551 | LOC_Os12g20220 |
| Translocation | Chr11 | 12942262 | Chr4 | 779243 | 2 |
| Translocation | Chr3 | 13539829 | Chr2 | 30014446 | LOC_Os03g23860 |
| Translocation | Chr9 | 16882224 | Chr2 | 34742587 | 2 |
| Translocation | Chr10 | 17232345 | Chr1 | 7861565 | |
| Translocation | Chr5 | 17883172 | Chr3 | 31648212 | |
| Translocation | Chr2 | 27133113 | Chr1 | 25821321 |
