Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2755-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2755-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 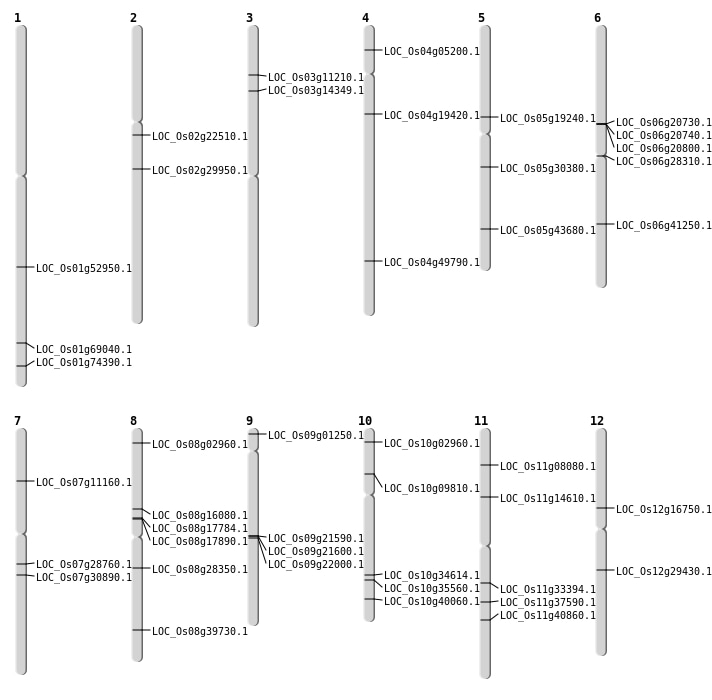
|
| Variant Information |
|---|
Single base substitutions: 68
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 21816863 | A-G | HOMO | INTRON | |
| SBS | Chr1 | 37507648 | G-C | HET | ||
| SBS | Chr1 | 37507649 | C-A | HET | ||
| SBS | Chr1 | 37817119 | T-C | HOMO | INTERGENIC | |
| SBS | Chr1 | 43089237 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g74390 |
| SBS | Chr1 | 43240515 | C-A | HET | INTERGENIC | |
| SBS | Chr1 | 9434029 | T-C | HET | ||
| SBS | Chr10 | 19019792 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g35560 |
| SBS | Chr10 | 19022677 | C-G | HET | INTRON | |
| SBS | Chr10 | 4405639 | G-A | HET | ||
| SBS | Chr11 | 10280648 | G-A | HOMO | INTRON | |
| SBS | Chr11 | 21218407 | T-A | HET | INTERGENIC | |
| SBS | Chr11 | 3427229 | C-T | HET | UTR_3_PRIME | |
| SBS | Chr11 | 4226652 | A-C | HET | NON_SYNONYMOUS_CODING | LOC_Os11g08080 |
| SBS | Chr12 | 13267692 | C-T | HET | ||
| SBS | Chr12 | 14987782 | G-A | HET | ||
| SBS | Chr12 | 22100654 | G-A | HET | ||
| SBS | Chr12 | 24232295 | T-A | HET | ||
| SBS | Chr12 | 8312897 | G-T | HET | INTERGENIC | |
| SBS | Chr12 | 9847784 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 13432238 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os02g22510 |
| SBS | Chr2 | 15002416 | C-T | HET | ||
| SBS | Chr2 | 17801513 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g29950 |
| SBS | Chr2 | 20841243 | A-C | Homo | ||
| SBS | Chr2 | 2634256 | G-A | Homo | ||
| SBS | Chr2 | 31025719 | T-C | HET | ||
| SBS | Chr2 | 33538703 | T-C | HET | ||
| SBS | Chr2 | 34470348 | C-T | HET | ||
| SBS | Chr2 | 35342528 | T-C | HET | INTRON | |
| SBS | Chr2 | 4687717 | A-T | HET | INTRON | |
| SBS | Chr3 | 12421208 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 13559330 | C-T | Homo | ||
| SBS | Chr3 | 16954453 | C-T | HET | ||
| SBS | Chr3 | 21377557 | C-T | HET | ||
| SBS | Chr3 | 22544820 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 23504046 | C-T | Homo | ||
| SBS | Chr3 | 28987007 | T-A | HET | INTERGENIC | |
| SBS | Chr3 | 29419488 | C-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 3746463 | G-A | HET | ||
| SBS | Chr3 | 7809773 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g14349 |
| SBS | Chr4 | 1733217 | A-G | HET | INTERGENIC | |
| SBS | Chr4 | 20648675 | G-T | HET | INTERGENIC | |
| SBS | Chr4 | 2596474 | G-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os04g05200 |
| SBS | Chr4 | 33051752 | C-T | HET | ||
| SBS | Chr4 | 34835738 | C-T | HET | ||
| SBS | Chr4 | 35149459 | C-A | HET | ||
| SBS | Chr5 | 11174840 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os05g19240 |
| SBS | Chr5 | 16446053 | C-T | HET | INTRON | |
| SBS | Chr5 | 20131613 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 7640675 | A-T | Homo | ||
| SBS | Chr6 | 20681932 | T-C | HET | ||
| SBS | Chr6 | 21705788 | C-T | HET | ||
| SBS | Chr6 | 22043188 | A-G | HOMO | INTERGENIC | |
| SBS | Chr6 | 25372163 | A-T | HET | ||
| SBS | Chr6 | 2740088 | C-T | HET | ||
| SBS | Chr6 | 8983079 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 18289125 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g30890 |
| SBS | Chr7 | 20419501 | T-C | HET | INTERGENIC | |
| SBS | Chr7 | 2042434 | G-T | Homo | ||
| SBS | Chr7 | 20923497 | C-T | Homo | ||
| SBS | Chr7 | 251638 | T-A | HET | INTERGENIC | |
| SBS | Chr7 | 2524882 | G-C | Homo | ||
| SBS | Chr7 | 5110292 | A-G | HET | ||
| SBS | Chr7 | 6910651 | T-C | HET | ||
| SBS | Chr7 | 8252066 | T-C | HET | INTERGENIC | |
| SBS | Chr8 | 10312430 | A-G | HET | ||
| SBS | Chr8 | 17769894 | A-G | HET | ||
| SBS | Chr8 | 25156066 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os08g39730 |
Deletions: 64
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr11 | 180941 | 180967 | 26 | |
| Deletion | Chr7 | 1040851 | 1040856 | 5 | |
| Deletion | Chr10 | 1220827 | 1220828 | 1 | LOC_Os10g02960 |
| Deletion | Chr8 | 1288043 | 1288059 | 16 | LOC_Os08g02960 |
| Deletion | Chr8 | 1362522 | 1362523 | 1 | |
| Deletion | Chr8 | 1421213 | 1421215 | 2 | |
| Deletion | Chr5 | 1572994 | 1572995 | 1 | |
| Deletion | Chr1 | 4072974 | 4072980 | 6 | |
| Deletion | Chr2 | 4406562 | 4406563 | 1 | |
| Deletion | Chr2 | 4883156 | 4883157 | 1 | |
| Deletion | Chr11 | 5168027 | 5168032 | 5 | |
| Deletion | Chr10 | 5305299 | 5305300 | 1 | LOC_Os10g09810 |
| Deletion | Chr7 | 6156198 | 6156199 | 1 | LOC_Os07g11160 |
| Deletion | Chr7 | 6227649 | 6227652 | 3 | |
| Deletion | Chr12 | 8068786 | 8068788 | 2 | |
| Deletion | Chr11 | 8233027 | 8233075 | 48 | LOC_Os11g14610 |
| Deletion | Chr8 | 8545561 | 8545562 | 1 | |
| Deletion | Chr5 | 8545926 | 8545927 | 1 | |
| Deletion | Chr10 | 8728114 | 8728115 | 1 | |
| Deletion | Chr1 | 9514665 | 9514667 | 2 | |
| Deletion | Chr12 | 9600340 | 9600341 | 1 | LOC_Os12g16750 |
| Deletion | Chr8 | 9807985 | 9807986 | 1 | LOC_Os08g16080 |
| Deletion | Chr5 | 9893210 | 9893211 | 1 | |
| Deletion | Chr7 | 9914851 | 9914852 | 1 | |
| Deletion | Chr1 | 10767259 | 10767269 | 10 | |
| Deletion | Chr8 | 10893001 | 10908000 | 14999 | 2 |
| Deletion | Chr4 | 11041854 | 11041856 | 2 | |
| Deletion | Chr12 | 11725122 | 11725136 | 14 | |
| Deletion | Chr6 | 11934001 | 11951000 | 16999 | 3 |
| Deletion | Chr1 | 12680300 | 12680306 | 6 | |
| Deletion | Chr9 | 13052001 | 13081000 | 28999 | 3 |
| Deletion | Chr9 | 13771289 | 13771316 | 27 | |
| Deletion | Chr8 | 14221350 | 14221351 | 1 | |
| Deletion | Chr11 | 14925126 | 14925127 | 1 | |
| Deletion | Chr11 | 15070869 | 15070888 | 19 | |
| Deletion | Chr6 | 16082829 | 16082830 | 1 | LOC_Os06g28310 |
| Deletion | Chr12 | 16674017 | 16674018 | 1 | |
| Deletion | Chr6 | 16785006 | 16785011 | 5 | |
| Deletion | Chr8 | 17445053 | 17445054 | 1 | |
| Deletion | Chr12 | 17479698 | 17479748 | 50 | LOC_Os12g29430 |
| Deletion | Chr6 | 17783796 | 17783797 | 1 | |
| Deletion | Chr5 | 18350712 | 18350713 | 1 | |
| Deletion | Chr11 | 18406151 | 18406152 | 1 | |
| Deletion | Chr3 | 18975401 | 18975402 | 1 | |
| Deletion | Chr3 | 19229638 | 19229642 | 4 | |
| Deletion | Chr4 | 19764151 | 19764152 | 1 | |
| Deletion | Chr1 | 20162089 | 20162104 | 15 | |
| Deletion | Chr5 | 20510461 | 20510462 | 1 | |
| Deletion | Chr5 | 21422062 | 21422064 | 2 | |
| Deletion | Chr10 | 21456586 | 21456591 | 5 | LOC_Os10g40060 |
| Deletion | Chr11 | 22189730 | 22189731 | 1 | LOC_Os11g37590 |
| Deletion | Chr2 | 22470466 | 22470474 | 8 | |
| Deletion | Chr1 | 22482043 | 22482051 | 8 | |
| Deletion | Chr12 | 22891456 | 22891457 | 1 | |
| Deletion | Chr3 | 25248173 | 25248174 | 1 | |
| Deletion | Chr11 | 25389394 | 25389396 | 2 | |
| Deletion | Chr5 | 25393618 | 25393627 | 9 | LOC_Os05g43680 |
| Deletion | Chr6 | 26250904 | 26250908 | 4 | |
| Deletion | Chr1 | 29569677 | 29569687 | 10 | |
| Deletion | Chr1 | 30443824 | 30443825 | 1 | LOC_Os01g52950 |
| Deletion | Chr2 | 30911870 | 30911871 | 1 | |
| Deletion | Chr1 | 31167182 | 31167188 | 6 | |
| Deletion | Chr1 | 34942764 | 34942767 | 3 | |
| Deletion | Chr1 | 36629159 | 36629160 | 1 |
Insertions: 25
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 4849324 | 4849327 | 4 | |
| Insertion | Chr1 | 7292911 | 7292911 | 1 | |
| Insertion | Chr10 | 5264583 | 5264583 | 1 | |
| Insertion | Chr12 | 22833330 | 22833331 | 2 | |
| Insertion | Chr12 | 25686369 | 25686369 | 1 | |
| Insertion | Chr12 | 4997260 | 4997273 | 14 | |
| Insertion | Chr12 | 6994745 | 6994745 | 1 | |
| Insertion | Chr2 | 32768240 | 32768240 | 1 | |
| Insertion | Chr2 | 8657225 | 8657226 | 2 | |
| Insertion | Chr3 | 22475639 | 22475639 | 1 | |
| Insertion | Chr3 | 31582364 | 31582365 | 2 | |
| Insertion | Chr4 | 10479946 | 10479946 | 1 | |
| Insertion | Chr4 | 6055173 | 6055176 | 4 | |
| Insertion | Chr5 | 11128475 | 11128475 | 1 | |
| Insertion | Chr5 | 17611700 | 17611700 | 1 | LOC_Os05g30380 |
| Insertion | Chr5 | 6652240 | 6652240 | 1 | |
| Insertion | Chr5 | 8994961 | 8994961 | 1 | |
| Insertion | Chr6 | 22684837 | 22684837 | 1 | |
| Insertion | Chr6 | 29983219 | 29983219 | 1 | |
| Insertion | Chr7 | 16836657 | 16836657 | 1 | LOC_Os07g28760 |
| Insertion | Chr7 | 27351637 | 27351637 | 1 | |
| Insertion | Chr8 | 15899208 | 15899208 | 1 | |
| Insertion | Chr9 | 11135138 | 11135138 | 1 | |
| Insertion | Chr9 | 18946833 | 18946833 | 1 | |
| Insertion | Chr9 | 214333 | 214333 | 1 | LOC_Os09g01250 |
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr4 | 9780788 | 9838284 | |
| Inversion | Chr4 | 10802700 | 10820855 | LOC_Os04g19420 |
| Inversion | Chr4 | 10802702 | 10820860 | LOC_Os04g19420 |
| Inversion | Chr11 | 18825586 | 18828949 | |
| Inversion | Chr6 | 24684474 | 24686396 | LOC_Os06g41250 |
Translocations: 30
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr5 | 987248 | Chr2 | 30921565 | |
| Translocation | Chr2 | 2071734 | Chr1 | 1499653 | |
| Translocation | Chr11 | 3129576 | Chr6 | 18189937 | |
| Translocation | Chr8 | 4126990 | Chr1 | 3219749 | |
| Translocation | Chr5 | 7147889 | Chr3 | 10130140 | |
| Translocation | Chr12 | 7165584 | Chr1 | 30931778 | |
| Translocation | Chr8 | 8178750 | Chr3 | 28268378 | |
| Translocation | Chr7 | 10123760 | Chr4 | 10495508 | |
| Translocation | Chr12 | 10782173 | Chr3 | 13483603 | |
| Translocation | Chr6 | 11735568 | Chr5 | 18456079 | |
| Translocation | Chr6 | 11736057 | Chr5 | 18456109 | |
| Translocation | Chr12 | 12183087 | Chr2 | 9649794 | |
| Translocation | Chr4 | 15031765 | Chr2 | 10522098 | |
| Translocation | Chr11 | 15249059 | Chr1 | 11769608 | |
| Translocation | Chr8 | 16449095 | Chr6 | 24693727 | |
| Translocation | Chr10 | 18451785 | Chr6 | 8105161 | LOC_Os10g34614 |
| Translocation | Chr11 | 18508356 | Chr1 | 40950870 | |
| Translocation | Chr8 | 18628532 | Chr4 | 2531450 | |
| Translocation | Chr11 | 19361833 | Chr2 | 7462906 | |
| Translocation | Chr11 | 19361845 | Chr2 | 7462449 | |
| Translocation | Chr8 | 19749963 | Chr2 | 5753914 | |
| Translocation | Chr11 | 19762910 | Chr8 | 17299594 | 2 |
| Translocation | Chr9 | 21485649 | Chr3 | 5772142 | LOC_Os03g11210 |
| Translocation | Chr9 | 21486962 | Chr3 | 5772135 | LOC_Os03g11210 |
| Translocation | Chr9 | 21486980 | Chr1 | 40120894 | LOC_Os01g69040 |
| Translocation | Chr9 | 21487539 | Chr1 | 40120848 | LOC_Os01g69040 |
| Translocation | Chr11 | 24445386 | Chr4 | 29686988 | 2 |
| Translocation | Chr11 | 24994391 | Chr4 | 17873076 | |
| Translocation | Chr6 | 25586141 | Chr2 | 15328433 | |
| Translocation | Chr11 | 25898149 | Chr9 | 4534242 |
