Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2781-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2781-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 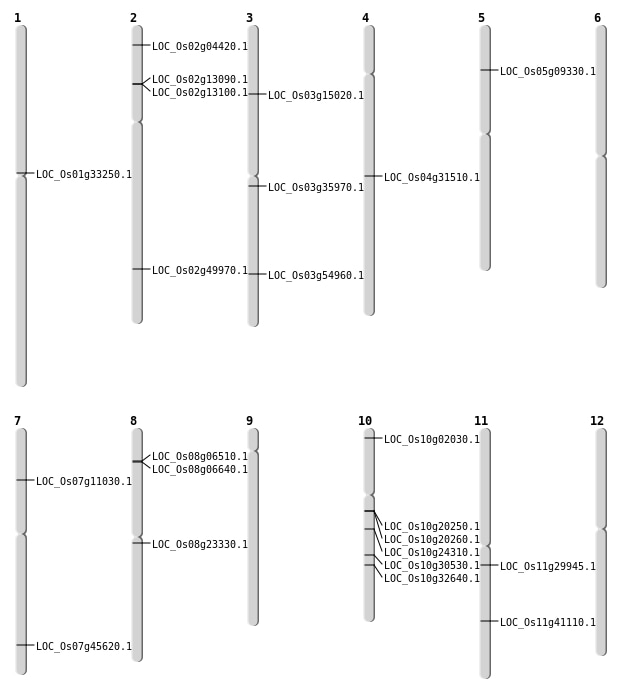
|
| Variant Information |
|---|
Single base substitutions: 77
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 10836186 | G-A | HET | ||
| SBS | Chr1 | 11991041 | G-T | HET | ||
| SBS | Chr1 | 13097391 | G-A | HET | ||
| SBS | Chr1 | 18313692 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g33250 |
| SBS | Chr1 | 18355731 | G-A | HET | ||
| SBS | Chr1 | 22206144 | G-A | Homo | ||
| SBS | Chr1 | 22206145 | C-A | Homo | ||
| SBS | Chr1 | 24297818 | G-A | HET | ||
| SBS | Chr1 | 27213181 | T-C | HET | ||
| SBS | Chr1 | 27213185 | G-A | HET | ||
| SBS | Chr1 | 27687661 | G-T | Homo | ||
| SBS | Chr1 | 35769529 | A-T | HET | ||
| SBS | Chr1 | 35769530 | A-T | HET | ||
| SBS | Chr1 | 3654108 | T-C | Homo | ||
| SBS | Chr10 | 12171661 | C-T | HET | ||
| SBS | Chr10 | 15876262 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os10g30530 |
| SBS | Chr10 | 15876264 | C-T | HET | ||
| SBS | Chr10 | 21518305 | G-C | HET | ||
| SBS | Chr10 | 296661 | A-T | HET | ||
| SBS | Chr10 | 3441925 | C-T | HET | ||
| SBS | Chr10 | 647647 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os10g02030 |
| SBS | Chr11 | 1471704 | T-C | HET | ||
| SBS | Chr11 | 19307765 | A-G | Homo | ||
| SBS | Chr11 | 19330822 | C-T | HET | ||
| SBS | Chr11 | 19603891 | G-A | HET | ||
| SBS | Chr11 | 23896656 | A-T | HET | ||
| SBS | Chr11 | 27682494 | G-A | HET | ||
| SBS | Chr12 | 1617182 | G-C | HET | ||
| SBS | Chr12 | 22592485 | G-A | HET | ||
| SBS | Chr2 | 11644451 | C-A | HET | ||
| SBS | Chr2 | 13004294 | G-C | HET | ||
| SBS | Chr2 | 17060839 | C-A | HET | ||
| SBS | Chr2 | 1948097 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g04420 |
| SBS | Chr2 | 1948098 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g04420 |
| SBS | Chr2 | 1948102 | G-A | HET | ||
| SBS | Chr2 | 2481741 | T-G | HET | ||
| SBS | Chr2 | 31284625 | C-T | HET | ||
| SBS | Chr2 | 31359046 | C-T | HET | ||
| SBS | Chr3 | 15523904 | C-T | Homo | ||
| SBS | Chr3 | 3099465 | A-T | Homo | ||
| SBS | Chr3 | 3099467 | A-T | Homo | ||
| SBS | Chr3 | 3099469 | C-G | Homo | ||
| SBS | Chr3 | 31241921 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g54960 |
| SBS | Chr3 | 5090404 | G-A | Homo | ||
| SBS | Chr3 | 7563654 | G-A | Homo | ||
| SBS | Chr4 | 15903436 | T-G | HET | ||
| SBS | Chr4 | 18840180 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os04g31510 |
| SBS | Chr4 | 20277944 | C-A | HET | ||
| SBS | Chr4 | 2412520 | G-A | HET | ||
| SBS | Chr4 | 24468499 | C-T | HET | ||
| SBS | Chr4 | 30391443 | G-T | HET | ||
| SBS | Chr4 | 31690447 | T-A | HET | ||
| SBS | Chr4 | 3865011 | C-A | HET | ||
| SBS | Chr4 | 8019300 | C-A | HET | ||
| SBS | Chr5 | 13565242 | A-C | Homo | ||
| SBS | Chr5 | 567094 | T-A | HET | ||
| SBS | Chr5 | 567095 | G-A | HET | ||
| SBS | Chr6 | 10568393 | G-A | HET | ||
| SBS | Chr6 | 19518495 | G-A | Homo | ||
| SBS | Chr6 | 20203526 | G-A | HET | ||
| SBS | Chr6 | 26767404 | T-A | Homo | ||
| SBS | Chr7 | 12108402 | C-A | HET | ||
| SBS | Chr7 | 12516782 | G-A | HET | ||
| SBS | Chr7 | 17710919 | G-T | HET | ||
| SBS | Chr7 | 6077022 | T-C | Homo | NON_SYNONYMOUS_CODING | LOC_Os07g11030 |
| SBS | Chr7 | 9497139 | C-A | HET | ||
| SBS | Chr8 | 3744851 | G-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os08g06640 |
| SBS | Chr8 | 5533356 | C-T | HET | ||
| SBS | Chr9 | 10094933 | C-T | HET | ||
| SBS | Chr9 | 10245726 | A-T | HET | ||
| SBS | Chr9 | 14956831 | C-T | HET | ||
| SBS | Chr9 | 15441555 | G-A | HET | ||
| SBS | Chr9 | 15728607 | T-C | HET | ||
| SBS | Chr9 | 17924429 | A-C | HET | ||
| SBS | Chr9 | 22689733 | C-T | HET | ||
| SBS | Chr9 | 4882231 | G-A | HET | ||
| SBS | Chr9 | 8796915 | A-G | HET |
Deletions: 28
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr4 | 448497 | 448515 | 18 | |
| Deletion | Chr8 | 1018493 | 1018511 | 18 | |
| Deletion | Chr8 | 2618543 | 2618547 | 4 | |
| Deletion | Chr3 | 2651551 | 2651555 | 4 | |
| Deletion | Chr3 | 2660065 | 2660073 | 8 | |
| Deletion | Chr8 | 3686664 | 3686680 | 16 | LOC_Os08g06510 |
| Deletion | Chr1 | 3741748 | 3741751 | 3 | |
| Deletion | Chr5 | 5032669 | 5032675 | 6 | |
| Deletion | Chr9 | 5281311 | 5281313 | 2 | |
| Deletion | Chr12 | 6920294 | 6920315 | 21 | |
| Deletion | Chr2 | 6926001 | 6960000 | 33999 | 2 |
| Deletion | Chr3 | 8198649 | 8198652 | 3 | LOC_Os03g15020 |
| Deletion | Chr10 | 10177001 | 10189000 | 11999 | 2 |
| Deletion | Chr2 | 10879663 | 10879669 | 6 | |
| Deletion | Chr2 | 11866050 | 11866062 | 12 | |
| Deletion | Chr1 | 15150457 | 15150458 | 1 | |
| Deletion | Chr4 | 16801537 | 16801538 | 1 | |
| Deletion | Chr10 | 17095616 | 17095626 | 10 | LOC_Os10g32640 |
| Deletion | Chr11 | 17403435 | 17403452 | 17 | LOC_Os11g29945 |
| Deletion | Chr10 | 18011496 | 18011515 | 19 | |
| Deletion | Chr4 | 18175694 | 18175695 | 1 | |
| Deletion | Chr11 | 19599648 | 19599652 | 4 | |
| Deletion | Chr3 | 19951681 | 19951682 | 1 | LOC_Os03g35970 |
| Deletion | Chr5 | 20080460 | 20080474 | 14 | |
| Deletion | Chr5 | 20105523 | 20105529 | 6 | |
| Deletion | Chr2 | 20112780 | 20112781 | 1 | |
| Deletion | Chr3 | 30098407 | 30098408 | 1 | |
| Deletion | Chr2 | 32724924 | 32724926 | 2 |
No Insertion
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr8 | 1344416 | 1446167 | |
| Inversion | Chr11 | 24631004 | 25164189 | LOC_Os11g41110 |
| Inversion | Chr11 | 24631045 | 25164229 | LOC_Os11g41110 |
| Inversion | Chr2 | 30392248 | 30529266 | 2 |
| Inversion | Chr2 | 30392252 | 30529265 | 2 |
Translocations: 8
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr8 | 1344407 | Chr4 | 30345387 | |
| Translocation | Chr8 | 1361488 | Chr2 | 2418050 | |
| Translocation | Chr9 | 4916918 | Chr2 | 11824568 | |
| Translocation | Chr10 | 10189304 | Chr4 | 14595105 | |
| Translocation | Chr10 | 12452443 | Chr7 | 27239357 | 2 |
| Translocation | Chr8 | 14086287 | Chr5 | 5212205 | 2 |
| Translocation | Chr12 | 18686678 | Chr5 | 13636098 | |
| Translocation | Chr7 | 27769966 | Chr1 | 29676763 |
