Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2883-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2883-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 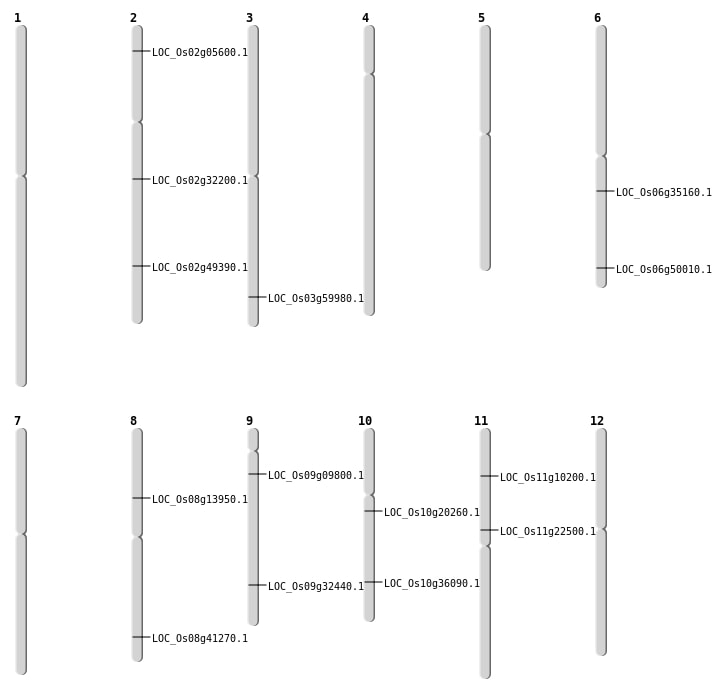
|
| Variant Information |
|---|
Single base substitutions: 44
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 14458287 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 22065252 | A-G | HOMO | INTERGENIC | |
| SBS | Chr1 | 38351088 | C-T | HOMO | INTERGENIC | |
| SBS | Chr10 | 15088133 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 16416194 | T-C | HET | INTERGENIC | |
| SBS | Chr10 | 3668606 | A-C | HET | INTERGENIC | |
| SBS | Chr10 | 3668608 | A-C | HET | INTERGENIC | |
| SBS | Chr10 | 6401309 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 11746812 | T-C | HET | INTRON | |
| SBS | Chr11 | 12935029 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os11g22500 |
| SBS | Chr11 | 14100823 | T-C | HET | INTERGENIC | |
| SBS | Chr11 | 20783228 | T-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 4456690 | A-C | HOMO | INTERGENIC | |
| SBS | Chr12 | 11500530 | A-G | HOMO | INTRON | |
| SBS | Chr12 | 13435261 | C-T | HOMO | INTRON | |
| SBS | Chr12 | 9793253 | G-A | HOMO | INTRON | |
| SBS | Chr12 | 9864751 | C-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 12124272 | C-G | HET | INTERGENIC | |
| SBS | Chr2 | 20054662 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 25727567 | A-T | HOMO | INTRON | |
| SBS | Chr2 | 26987485 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 2723665 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os02g05600 |
| SBS | Chr2 | 30176294 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os02g49390 |
| SBS | Chr2 | 8240152 | C-A | HET | INTERGENIC | |
| SBS | Chr2 | 9331009 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 34121030 | C-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os03g59980 |
| SBS | Chr3 | 4711406 | T-C | HOMO | INTERGENIC | |
| SBS | Chr4 | 1206266 | C-G | HET | INTERGENIC | |
| SBS | Chr4 | 13378724 | C-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr4 | 13888214 | G-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 17394803 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 27441010 | G-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 1965199 | A-G | HOMO | INTERGENIC | |
| SBS | Chr5 | 20672718 | C-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 20860796 | C-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 9425717 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 20491399 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os06g35160 |
| SBS | Chr6 | 30285103 | G-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os06g50010 |
| SBS | Chr6 | 30285104 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os06g50010 |
| SBS | Chr6 | 94616 | A-T | HET | INTERGENIC | |
| SBS | Chr8 | 26062588 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os08g41270 |
| SBS | Chr8 | 27801040 | A-C | HET | INTRON | |
| SBS | Chr8 | 8359394 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os08g13950 |
| SBS | Chr9 | 5313975 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os09g09800 |
Deletions: 22
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr2 | 1504979 | 1504980 | 1 | |
| Deletion | Chr9 | 2623773 | 2623828 | 55 | |
| Deletion | Chr10 | 3457814 | 3457817 | 3 | |
| Deletion | Chr11 | 5533540 | 5533549 | 9 | LOC_Os11g10200 |
| Deletion | Chr10 | 6377357 | 6377365 | 8 | |
| Deletion | Chr7 | 7250670 | 7250691 | 21 | |
| Deletion | Chr1 | 7806665 | 7806681 | 16 | |
| Deletion | Chr7 | 7852045 | 7852048 | 3 | |
| Deletion | Chr10 | 10187577 | 10187578 | 1 | LOC_Os10g20260 |
| Deletion | Chr6 | 11961524 | 11961525 | 1 | |
| Deletion | Chr6 | 12916374 | 12916386 | 12 | |
| Deletion | Chr8 | 16441084 | 16441161 | 77 | |
| Deletion | Chr2 | 17171550 | 17171559 | 9 | |
| Deletion | Chr11 | 17184178 | 17184180 | 2 | |
| Deletion | Chr10 | 19299744 | 19299745 | 1 | LOC_Os10g36090 |
| Deletion | Chr9 | 19366599 | 19366602 | 3 | LOC_Os09g32440 |
| Deletion | Chr9 | 21123409 | 21123431 | 22 | |
| Deletion | Chr2 | 24224134 | 24224135 | 1 | |
| Deletion | Chr1 | 27462752 | 27462753 | 1 | |
| Deletion | Chr3 | 31917782 | 31917783 | 1 | |
| Deletion | Chr3 | 32505819 | 32505823 | 4 | |
| Deletion | Chr1 | 40965392 | 40965393 | 1 |
Insertions: 10
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr10 | 18648861 | 18899542 |
Translocations: 6
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr8 | 2233762 | Chr5 | 28810789 | |
| Translocation | Chr8 | 9335286 | Chr2 | 19023541 | LOC_Os02g32200 |
| Translocation | Chr10 | 18648841 | Chr1 | 31373321 | |
| Translocation | Chr10 | 18899533 | Chr1 | 31374973 | |
| Translocation | Chr11 | 22562642 | Chr10 | 9197237 | |
| Translocation | Chr11 | 22562649 | Chr10 | 9196660 |
